| Type: | Package |
| Title: | Matrix Functions for Teaching and Learning Linear Algebra and Multivariate Statistics |
| Version: | 1.0.1 |
| Date: | 2025-10-04 |
| Maintainer: | Michael Friendly <friendly@yorku.ca> |
| Description: | A collection of matrix functions for teaching and learning matrix linear algebra as used in multivariate statistical methods. Many of these functions are designed for tutorial purposes in learning matrix algebra ideas using R. In some cases, functions are provided for concepts available elsewhere in R, but where the function call or name is not obvious. In other cases, functions are provided to show or demonstrate an algorithm. In addition, a collection of functions are provided for drawing vector diagrams in 2D and 3D and for rendering matrix expressions and equations in LaTeX. |
| License: | GPL-2 | GPL-3 [expanded from: GPL (≥ 2)] |
| Language: | en-US |
| URL: | https://github.com/friendly/matlib, http://friendly.github.io/matlib/ |
| BugReports: | https://github.com/friendly/matlib/issues |
| LazyData: | TRUE |
| Depends: | R (≥ 4.2.0) |
| Suggests: | carData, webshot2, markdown, bookdown, clipr |
| Imports: | xtable, MASS, rgl, car, methods, dplyr, knitr, rmarkdown, rstudioapi |
| VignetteBuilder: | knitr |
| RoxygenNote: | 7.3.2 |
| Encoding: | UTF-8 |
| NeedsCompilation: | no |
| Packaged: | 2025-10-05 14:22:17 UTC; friendly |
| Author: | Michael Friendly |
| Repository: | CRAN |
| Date/Publication: | 2025-10-09 09:50:02 UTC |
matlib: Matrix Functions for Teaching and Learning Linear Algebra and Multivariate Statistics.
Description
These functions are designed mainly for tutorial purposes in teaching & learning matrix algebra ideas and applications to statistical methods using R.
Details
In some cases, functions are provided for concepts available
elsewhere in R, but where the function call or name is not obvious. In other
cases, functions are provided to show or demonstrate an algorithm, sometimes
providing a verbose argument to print the details of computations.
In addition, a collection of functions are provided for drawing vector diagrams in 2D and 3D.
These are not meant for production uses. Other methods are more efficient for larger problems.
Topics
The functions in this package are grouped under the following topics
Convenience functions:
tr,R,J,len,vec,Proj,mpower,vandermodeDeterminants: functions for calculating determinants by cofactor expansion
minor,cofactor,rowMinors,rowCofactorsElementary row operations: functions for solving linear equations "manually" by the steps used in row echelon form and Gaussian elimination
rowadd,rowmult,rowswapLinear equations: functions to illustrate linear equations of the form $A x = b$
showEqn,plotEqnGaussian elimination: functions for illustrating Gaussian elimination for solving systems of linear equations of the form $A x = b$.
gaussianElimination,Inverse,inv,echelon,Ginv,LU,cholesky,swpEigenvalues: functions to illustrate the algorithms for calculating eigenvalues and eigenvectors
eigen,SVD,powerMethod,showEigVector diagrams: functions for drawing vector diagrams in 2D and 3D
arrows3d,corner,arc,pointOnLine,vectors,vectors3d,regvec3d
Most of these ideas and implementations arose in courses and books by the authors. [Psychology 6140](http://friendly.apps01.yorku.ca/psy6140/) was a starting point. Fox (1984) introduced illustrations of vector geometry.
macOS Installation Note
The functions that draw 3D graphs use the rgl package. On macOS, the rgl package requires that XQuartz be installed. After installing XQuartz, it's necessary either to log out of and back into your macOS account or to reboot your Mac.
References
Fox, J. Linear Statistical Models and Related Methods. John Wiley and Sons, 1984
Fox, J. and Friendly, M. (2016). "Visualizing Simultaneous Linear Equations, Geometric Vectors, and Least-Squares Regression with the matlib Package for R". useR Conference, Stanford, CA, June 27 - June 30, 2016.
Determinant of a Square Matrix
Description
Returns the determinant of a square matrix X,
computed either by Gaussian elimination, expansion by cofactors, or as the product of the eigenvalues of the matrix.
If the latter, X must be symmetric.
Usage
Det(
X,
method = c("elimination", "eigenvalues", "cofactors"),
verbose = FALSE,
fractions = FALSE,
...
)
Arguments
X |
a square matrix |
method |
one of '"elimination"' (the default), '"eigenvalues"', or '"cofactors"' (for computation by minors and cofactors) |
verbose |
logical; if |
fractions |
logical; if |
... |
arguments passed to |
Value
the determinant of X
Author(s)
John Fox
See Also
det for the base R function
Other determinants:
adjoint(),
cofactor(),
minor(),
rowCofactors(),
rowMinors()
Examples
A <- matrix(c(1,2,3,2,5,6,3,6,10), 3, 3) # nonsingular, symmetric
A
Det(A)
Det(A, verbose=TRUE, fractions=TRUE)
B <- matrix(1:9, 3, 3) # a singular matrix
B
Det(B)
C <- matrix(c(1, .5, .5, 1), 2, 2) # square, symmetric, nonsingular
Det(C)
Det(C, method="eigenvalues")
Det(C, method="cofactors")
Eigen Decomposition of a Square Symmetric Matrix
Description
Eigen calculates the eigenvalues and eigenvectors of a square, symmetric matrix using the iterated QR decomposition
Usage
Eigen(X, tol = sqrt(.Machine$double.eps), max.iter = 100, retain.zeroes = TRUE)
Arguments
X |
a square symmetric matrix |
tol |
tolerance passed to |
max.iter |
maximum number of QR iterations |
retain.zeroes |
logical; retain 0 eigenvalues? |
Value
a list of two elements: values– eigenvalues, vectors– eigenvectors
Author(s)
John Fox and Georges Monette
See Also
Examples
C <- matrix(c(1,2,3,2,5,6,3,6,10), 3, 3) # nonsingular, symmetric
C
EC <- Eigen(C) # eigenanalysis of C
EC$vectors %*% diag(EC$values) %*% t(EC$vectors) # check
Create a LaTeX Equation Wrapper
Description
The Eqn function is designed to produce LaTeX expressions of mathematical
equations for writing.
The output can be copied/pasted into documents or
used directly in chunks in .Rmd, .Rnw, or .qmd
documents to compile to equations.
Eqn wraps the equations generated by its arguments
in either a \begin{equation} ...\end{equation} or
\begin{align} ...\end{align} LaTeX environment. See also
ref for consistent inline referencing of numbered equations in documents.
In a code chunk, use the chunk options results='asis', echo=FALSE to show only
the result of compiling the LaTeX expressions. For example,
```{r results = "asis", echo = FALSE}
Eqn("\mathbf{X} = \mathbf{U} \mathbf{\Lambda} \mathbf{V}", label='eq:svd')
```
Note that you can avoid the "leaning toothpick syndrome" of all those doubled backslashes by using R's new (as of 4.0.0)
"raw strings", composed as r"(...)" or r"{...}"
```{r results = "asis", echo = FALSE}
Eqn(r"{\mathbf{X} = \mathbf{U} \mathbf{\Lambda} \mathbf{V}}", label = 'eq:svn')
```
A collection of helper functions, such as Eqn_newline, Eqn_hspace
facilitate formatting of equations and functions like Eqn_overset and Eqn_overbrace
provide for decorators over or under a LaTeX expression or matrix. See Eqn_helpers
for their descriptions and examples.
ref{} provides inline references to equations in R
markdown and Quarto documents.
Depending on the output type this function will provide the correct
inline wrapper for MathJax or LaTeX equations. This provides more
consistent referencing when switching between HTML and PDF outputs as
well as documentation types (e.g., .Rmd vs .qmd).
Usage
Eqn(
...,
label = NULL,
align = FALSE,
preview = getOption("previewEqn"),
html_output = knitr::is_html_output(),
quarto = getOption("quartoEqn"),
mat_args = list(),
preview.pdf = FALSE,
preview.packages = NULL
)
ref(
label,
parentheses = TRUE,
html_output = knitr::is_html_output(),
quarto = getOption("quartoEqn")
)
Arguments
... |
comma separated LaTeX expressions that are either a) a Note that user defined functions that use |
label |
character vector specifying the label to use (e.g., For compiled documents if an HTML output is detected (see |
align |
logical; use the |
preview |
logical; render an HTML version of the equation and display? This is intended for
testing purposes and is only applicable to interactive R sessions, though
for code testing purposes can be set globally
via |
html_output |
logical; use labels for HTML outputs instead of the LaTeX? Automatically
changed for compiled documents that support |
quarto |
logical; use Quarto referencing syntax? When |
mat_args |
list of arguments to be passed to |
preview.pdf |
logical; build a PDF of the preview equation? Generally not require unless additional LaTeX packages are required that are not supported by MathJax |
preview.packages |
character vector for adding additional LaTeX package information to the
equation preview. Only used when |
parentheses |
logical; include parentheses around the referenced equation? |
Author(s)
Phil Chalmers
References
Josiah Parry, Raw strings in R, https://josiahparry.com/posts/2023-01-19-raw-strings-in-r.html
See Also
Eqn_helpers, latexMatrix, matrix2latex, ref
Examples
# character input
Eqn('e=mc^2')
# show only the LaTeX code
Eqn('e=mc^2', preview=FALSE)
# Equation numbers & labels
Eqn('e=mc^2', label = 'eq:einstein')
Eqn("X=U \\lambda V", label='eq:svd')
# html_output and quarto outputs only show code
# (both auto detected in compiled documents)
Eqn('e=mc^2', label = 'eq:einstein', html_output = TRUE)
# Quarto output
Eqn('e=mc^2', label = 'eq-einstein', quarto = TRUE)
## Not run:
# The following requires LaTeX compilers to be pre-installed
# View PDF instead of HTML
Eqn('e=mc^2', preview.pdf=TRUE)
# Add extra LaTeX dependencies for PDF build
Eqn('\\bm{e}=mc^2', preview.pdf=TRUE,
preview.packages=c('amsmath', 'bm'))
## End(Not run)
# Multiple expressions
Eqn("e=mc^2",
Eqn_newline(),
"X=U \\lambda V", label='eq:svd')
# expressions that use cat() within their calls
Eqn('SVD = ',
latexMatrix("u", "n", "k"),
latexMatrix("\\lambda", "k", "k", diag=TRUE),
latexMatrix("v", "k", "p", transpose = TRUE),
label='eq:svd')
# align equations using & operator
Eqn("X &= U \\lambda V", Eqn_newline(),
"& = ", latexMatrix("u", "n", "k"),
latexMatrix("\\lambda", "k", "k", diag=TRUE),
latexMatrix("v", "k", "p", transpose = TRUE),
align=TRUE)
# numeric/character matrix example
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b <- matrix(c(8, -11, -3))
# numeric matrix wrapped internally
cbind(A,b) |> Eqn()
cbind(A,b) |> latexMatrix() |> Eqn()
# change numeric matrix brackets globally
cbind(A,b) |> Eqn(mat_args=list(matrix='bmatrix'))
# greater flexibility when using latexMatrix()
cbind(A, b) |> latexMatrix() |> partition(columns=3) |> Eqn()
# with showEqn()
showEqn(A, b, latex=TRUE) |> Eqn()
# used inside of Eqn() or manually defined labels in the document
Eqn('e = mc^2', label='eq:einstein')
# use within inline block via `r ref()`
ref('eq:einstein')
ref('eq:einstein', parentheses=FALSE)
ref('eq:einstein', html_output=TRUE)
# With Quarto
Eqn('e = mc^2', label='eq-einstein', quarto=TRUE)
ref('eq:einstein', quarto=TRUE)
ref('eq:einstein', quarto=TRUE, parentheses=FALSE)
Helpers to Compose Equations with Eqn
Description
These functions extend Eqn to facilitate composing LaTeX equations with desirable features
and pleasant typography.
Eqn_oversetandEqn_undersettypesets a label over or under a LaTeX expression or a"latexMatrix"objectEqn_overbraceandEqn_underbracetypesets a brace, with an optional label over or under an objectEqn_newline,Eqn_hspaceandEqn_vspacefacilitate spacing of parts of an equation.Eqn_sizechanges size of LaTeX text;Eqn_textincludes a literal string in equations.
Each of these (except Eqn_text) have aliases without the Eqn_ prefix for brevity.
For example, given the matrix A = matrix(1:4), 2, 2), the call Eqn(overset(A, "A"))
generates:
\overset{\mathbf{A}}
{ \begin{pmatrix}
1 & 3 \
2 & 4 \
\end{pmatrix}
}
When rendered in LaTeX, this produces:
\overset{\mathbf{A}}
{ \begin{pmatrix}
1 & 3 \\
2 & 4 \\
\end{pmatrix}
}
You can also use these for straight LaTeX expressions, such this equation showing and labeling
the Hat matrix in regression. See the examples for the call to underbrace for this.
\mathbf{\hat{y}} =
\underbrace{\mathbf{X}(\mathbf{X}^{\top}\mathbf{X})^{-1}
\mathbf{X}^{\top}}_{\mathbf{\mathbf{H}}}\mathbf{y}
Eqn_newline() emits a newline (\) in an equation, with
an optional increase to the padding following the newline.
Eqn_text() inserts a literal string to be rendered in a text font in an equation.
Eqn_hspace() is used to create (symmetric) equation spaces, most typically around
= signs
Input to lhs, rhs can be a
numeric to increase the size of the space or a
character vector to be passed to the LaTeX macro \hspace{}.
Eqn_vspace() inserts vertical space between lines in an equation.
Typically used for aligned, multiline equations.
Eqn_size() is used to increase or decrease the size of LaTeX text and equations. Can be applied
to a specific string or applied to all subsequent text until overwritten.
Usage
overset(
x,
label,
label.style = c("mathbf", "mathrm", "mathit", "mathsf", "mathcal", "mathtt", " ")
)
underset(
x,
label,
label.style = c("mathbf", "mathrm", "mathit", "mathsf", "mathcal", "mathtt", " ")
)
overbrace(
x,
label = NULL,
label.style = c("mathbf", "mathrm", "mathit", "mathsf", "mathcal", "mathtt", " ")
)
underbrace(
x,
label = NULL,
label.style = c("mathbf", "mathrm", "mathit", "mathsf", "mathcal", "mathtt", " ")
)
Eqn_overset(
x,
label,
label.style = c("mathbf", "mathrm", "mathit", "mathsf", "mathcal", "mathtt", " ")
)
Eqn_underset(
x,
label,
label.style = c("mathbf", "mathrm", "mathit", "mathsf", "mathcal", "mathtt", " ")
)
Eqn_overbrace(
x,
label = NULL,
label.style = c("mathbf", "mathrm", "mathit", "mathsf", "mathcal", "mathtt", " ")
)
Eqn_underbrace(
x,
label = NULL,
label.style = c("mathbf", "mathrm", "mathit", "mathsf", "mathcal", "mathtt", " ")
)
Eqn_newline(space = 0)
newline(space = 0)
Eqn_text(text)
Eqn_hspace(lhs = 5, mid = "", rhs = NULL, times = 1)
hspace(lhs = 5, mid = "", rhs = NULL, times = 1)
Eqn_vspace(space)
vspace(space)
Eqn_size(string, size = 0)
size(string, size = 0)
Arguments
x |
a numeric or character matrix, or a character string LaTeX expression or
a |
label |
a character string used as the label above or below the object |
label.style |
The name of a math font used to to typeset the label. One of
|
space |
includes extra vertical space. Metric of the vertical space must be 'ex', 'pt', 'mm', 'cm', 'em', 'bp', 'dd', 'pc', or 'in' |
text |
argument to be used within |
lhs |
spacing size. Can be a number between -1 and 6. -1 provides negative
spaces and 0 gives no spacing. Input can also be a character vector, which will be
passed to |
mid |
character vector to place in the middle of the space specification. Most
commonly this will be operators like |
rhs |
see lhs for details. If left as |
times |
number of times to repeat the spacings |
string |
a string that should have its text size modified. If missing
the size modifier is returned, which applies the size modifier
to the remainder of the text until reset with |
size |
numeric size of LaTeX text modifier,
ranging from -3 ( |
Value
Returns a character vector containing the LaTeX expressions for the given operation. You can pass
this to cat to display the result on the console, or include it inside a call
to Eqn to typeset it.
Author(s)
Michael Friendly, Phil Chalmers
Examples
library(matlib)
A <- matrix(1:4, 2, 2)
B <- matrix(4:1, 2, 2)
AB <- A + B
Eqn(overset(A, "A"))
# missing label: uses the name of the object
Eqn(overset(A))
# test just a character LaTeX expression
Eqn('a', overset('=', '?'), 'b')
# a labelled latexMatrix equation
Eqn(overset(A, "A"), "+",
overset(B, "B"), "=",
underset(AB, "A+B"))
# using a LaTeX expression as the label
Lambda <- latexMatrix("\\lambda", nrow=2, ncol=2,
diag=TRUE)
Eqn(overset(Lambda, "\\Lambda"))
# generate LaTeX expression for the Hat matrix, label as "H"
H <- "\\mathbf{X} (\\mathbf{X}^{\\top}\\mathbf{X})^{-1} \\mathbf{X}^{\\top}"
Eqn("\\mathbf{\\hat{y}} =", underbrace(H, "\\mathbf{H}"), "\\mathbf{y}")
# Combine this with overbrace
Eqn(overbrace(underbrace(H, "\\mathbf{H}"), "\\LARGE\\mathbf{\\hat{y}}"))
Eqn_newline()
Eqn_newline('10ex')
# more complete example
Eqn(underset("\\mathbf{X}", "(4 \\times 3)"),
"& = \\mathbf{U} \\mathbf{\\Lambda} \\mathbf{V}^\\top",
Eqn_newline('1ex'),
' & =',
latexMatrix("u", 4, 3),
latexMatrix("\\lambda", 3, 3, diag=TRUE),
latexMatrix("v", 3, 3, transpose = TRUE),
align=TRUE)
Eqn_hspace()
Eqn_hspace(3) # smaller
Eqn_hspace(3, times=2)
Eqn_hspace('1cm')
# symmetric spacing around mid
Eqn_hspace(mid='=')
Eqn_hspace(mid='=', times=2)
Eqn_vspace('1.5ex')
Eqn_vspace('1cm')
# set size globally
Eqn_size(size=3)
Eqn_size() # reset
# locally for defined string
string <- 'e = mc^2'
Eqn_size(string, size=1)
Generalized Inverse of a Matrix
Description
Ginv returns an arbitrary generalized inverse of the matrix A, using gaussianElimination.
Usage
Ginv(A, tol = sqrt(.Machine$double.eps), verbose = FALSE, fractions = FALSE)
Arguments
A |
numerical matrix |
tol |
tolerance for checking for 0 pivot |
verbose |
logical; if |
fractions |
logical; if |
Details
A generalized inverse is a matrix \mathbf{A}^- satisfying \mathbf{A A^- A} = \mathbf{A}.
The purpose of this function is mainly to show how the generalized inverse can be computed using Gaussian elimination.
Value
the generalized inverse of A, expressed as fractions if fractions=TRUE, or rounded
Author(s)
John Fox
See Also
ginv for a more generally usable function
Examples
A <- matrix(c(1,2,3,4,5,6,7,8,10), 3, 3) # a nonsingular matrix
A
Ginv(A, fractions=TRUE) # a generalized inverse of A = inverse of A
round(Ginv(A) %*% A, 6) # check
B <- matrix(1:9, 3, 3) # a singular matrix
B
Ginv(B, fractions=TRUE) # a generalized inverse of B
B %*% Ginv(B) %*% B # check
Gram-Schmidt Orthogonalization of a Matrix
Description
Carries out simple Gram-Schmidt orthogonalization of a matrix.
Treating the columns of the matrix X in the given order,
each successive column after the first is made orthogonal to all
previous columns by subtracting their projections on the current
column.
Usage
GramSchmidt(
X,
normalize = TRUE,
verbose = FALSE,
tol = sqrt(.Machine$double.eps),
omit_zero_columns = TRUE
)
Arguments
X |
a matrix |
normalize |
logical; should the resulting columns be normalized to unit length? The default is |
verbose |
logical; if |
tol |
the tolerance for detecting linear dependencies in the columns of a. The default is |
omit_zero_columns |
if |
Value
A matrix of the same size as X, with orthogonal columns (but with 0 columns removed by default)
Author(s)
Phil Chalmers, John Fox
Examples
(xx <- matrix(c( 1:3, 3:1, 1, 0, -2), 3, 3))
crossprod(xx)
(zz <- GramSchmidt(xx, normalize=FALSE))
zapsmall(crossprod(zz))
# normalized
(zz <- GramSchmidt(xx))
zapsmall(crossprod(zz))
# print steps
GramSchmidt(xx, verbose=TRUE)
# A non-invertible matrix; hence, it is of deficient rank
(xx <- matrix(c( 1:3, 3:1, 1, 0, -1), 3, 3))
R(xx)
crossprod(xx)
# GramSchmidt finds an orthonormal basis
(zz <- GramSchmidt(xx))
zapsmall(crossprod(zz))
Inverse of a Matrix
Description
Uses gaussianElimination to find the inverse of a square, non-singular matrix, X.
Usage
Inverse(X, tol = sqrt(.Machine$double.eps), verbose = FALSE, ...)
Arguments
X |
a square numeric matrix |
tol |
tolerance for checking for 0 pivot |
verbose |
logical; if |
... |
other arguments passed on |
Details
The method is purely didactic: The identity matrix, I, is appended to X, giving
X | I. Applying Gaussian elimination gives I | X^{-1}, and the portion corresponding
to X^{-1} is returned.
Value
the inverse of X
Author(s)
John Fox
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
Inverse(A)
Inverse(A, verbose=TRUE, fractions=TRUE)
Create a vector, matrix or array of constants
Description
This function creates a vector, matrix or array of constants, typically used for the unit vector or unit matrix in matrix expressions.
Usage
J(..., constant = 1, dimnames = NULL)
Arguments
... |
One or more arguments supplying the dimensions of the array, all non-negative integers |
constant |
The value of the constant used in the array |
dimnames |
Either |
Details
The "dimnames" attribute is optional: if present it is a list with one component for each dimension,
either NULL or a character vector of the length given by the element of the "dim" attribute for that
dimension. The list can be named, and the list names will be used as names for the dimensions.
Examples
J(3)
J(2,3)
J(2,3,2)
J(2,3, constant=2, dimnames=list(letters[1:2], LETTERS[1:3]))
X <- matrix(1:6, nrow=2, ncol=3)
dimnames(X) <- list(sex=c("M", "F"), day=c("Mon", "Wed", "Fri"))
J(2) %*% X # column sums
X %*% J(3) # row sums
LU Decomposition
Description
LU computes the LU decomposition of a matrix, A, such that P A = L U,
where L is a lower triangle matrix, U is an upper triangle, and P is a
permutation matrix.
Usage
LU(A, b, tol = sqrt(.Machine$double.eps), verbose = FALSE, ...)
Arguments
A |
coefficient matrix |
b |
right-hand side vector. When supplied the returned object will also contain the solved
|
tol |
tolerance for checking for 0 pivot |
verbose |
logical; if |
... |
additional arguments passed to |
Details
The LU decomposition is used to solve the equation A x = b by calculating
L(Ux - d) = 0, where Ld = b. If row exchanges are necessary for
A then the permutation matrix P will be required to exchange the rows in A;
otherwise, P will be an identity matrix and the LU equation will be simplified to
A = L U.
Value
A list of matrix components of the solution, P, L and U. If b
is supplied, the vectors d and x are also returned.
Author(s)
Phil Chalmers
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b <- c(8, -11, -3)
(ret <- LU(A)) # P is an identity; no row swapping
with(ret, L %*% U) # check that A = L * U
LU(A, b)
LU(A, b, verbose=TRUE)
LU(A, b, verbose=TRUE, fractions=TRUE)
# permutations required in this example
A <- matrix(c(1, 1, -1,
2, 2, 4,
1, -1, 1), 3, 3, byrow=TRUE)
b <- c(1, 2, 9)
(ret <- LU(A, b))
with(ret, P %*% A)
with(ret, L %*% U)
Moore-Penrose inverse of a matrix
Description
The Moore-Penrose inverse is a generalization of the regular inverse of a square, non-singular, symmetric matrix to other cases (rectangular, singular), yet retain similar properties to a regular inverse.
Usage
MoorePenrose(X, tol = sqrt(.Machine$double.eps))
Arguments
X |
A numeric matrix |
tol |
Tolerance for a singular (rank-deficient) matrix |
Value
The Moore-Penrose inverse of X
Examples
X <- matrix(rnorm(20), ncol=2)
# introduce a linear dependency in X[,3]
X <- cbind(X, 1.5*X[, 1] - pi*X[, 2])
Y <- MoorePenrose(X)
# demonstrate some properties of the M-P inverse
# X Y X = X
round(X %*% Y %*% X - X, 8)
# Y X Y = Y
round(Y %*% X %*% Y - Y, 8)
# X Y = t(X Y)
round(X %*% Y - t(X %*% Y), 8)
# Y X = t(Y X)
round(Y %*% X - t(Y %*% X), 8)
Projection of Vector y on columns of X
Description
Fitting a linear model, lm(y ~ X), by least squares can be thought of geometrically as the orthogonal projection of
y on the column space of X. This function is designed to allow exploration of projections
and orthogonality.
Usage
Proj(y, X, list = FALSE)
Arguments
y |
a vector, treated as a one-column matrix |
X |
a vector or matrix. Number of rows of |
list |
logical; if FALSE, return just the projected vector; otherwise returns a list |
Details
The projection is defined as P y where P = X (X'X)^- X'
and X^- is a generalized inverse.
Value
the projection of y on X (if list=FALSE) or a list with elements y and P
Author(s)
Michael Friendly
See Also
Other vector diagrams:
arc(),
arrows3d(),
circle3d(),
corner(),
plot.regvec3d(),
pointOnLine(),
regvec3d(),
vectors(),
vectors3d()
Examples
X <- matrix( c(1, 1, 1, 1, 1, -1, 1, -1), 4,2, byrow=TRUE)
y <- 1:4
Proj(y, X[,1]) # project y on unit vector
Proj(y, X[,2])
Proj(y, X)
# project unit vector on line between two points
y <- c(1,1)
p1 <- c(0,0)
p2 <- c(1,0)
Proj(y, cbind(p1, p2))
# orthogonal complements
y <- 1:4
yp <-Proj(y, X, list=TRUE)
yp$y
P <- yp$P
IP <- diag(4) - P
yc <- c(IP %*% y)
crossprod(yp$y, yc)
# P is idempotent: P P = P
P %*% P
all.equal(P, P %*% P)
QR Decomposition by Graham-Schmidt Orthonormalization
Description
QR computes the QR decomposition of a matrix, X, that is an orthonormal matrix, Q and an upper triangular
matrix, R, such that X = Q R.
Usage
QR(X, tol = sqrt(.Machine$double.eps))
Arguments
X |
a numeric matrix |
tol |
tolerance for detecting linear dependencies in the columns of |
Details
The QR decomposition plays an important role in many statistical techniques.
In particular it can be used to solve the equation Ax = b for given matrix A and vector b.
The function is included here simply to show the algorithm of Gram-Schmidt orthogonalization. The standard
qr function is faster and more accurate.
Value
a list of three elements, consisting of an orthonormal matrix Q, an upper triangular matrix R, and the rank
of the matrix X
Author(s)
John Fox and Georges Monette
See Also
Examples
A <- matrix(c(1,2,3,4,5,6,7,8,10), 3, 3) # a square nonsingular matrix
res <- QR(A)
res
q <- res$Q
zapsmall( t(q) %*% q) # check that q' q = I
r <- res$R
q %*% r # check that q r = A
# relation to determinant: det(A) = prod(diag(R))
det(A)
prod(diag(r))
B <- matrix(1:9, 3, 3) # a singular matrix
QR(B)
Rank of a Matrix
Description
Returns the rank of a matrix X, using the QR decomposition, QR.
Included here as a simple function, because rank does something different
and it is not obvious what to use for matrix rank.
Usage
R(X)
Arguments
X |
a matrix |
Value
rank of X
See Also
Examples
M <- outer(1:3, 3:1)
M
R(M)
M <- matrix(1:9, 3, 3)
M
R(M)
# why rank=2?
echelon(M)
set.seed(1234)
M <- matrix(sample(1:9), 3, 3)
M
R(M)
Singular Value Decomposition of a Matrix
Description
Compute the singular-value decomposition of a matrix X either by Jacobi
rotations (the default) or from the eigenstructure of X'X using
Eigen. Both methods are iterative.
The result consists of two orthonormal matrices, U, and V and the vector d
of singular values, such that X = U diag(d) V'.
Usage
SVD(
X,
method = c("Jacobi", "eigen"),
tol = sqrt(.Machine$double.eps),
max.iter = 100
)
Arguments
X |
a square symmetric matrix |
method |
either |
tol |
zero and convergence tolerance |
max.iter |
maximum number of iterations |
Details
The default method is more numerically stable, but the eigenstructure method is much simpler. Singular values of zero are not retained in the solution.
Value
a list of three elements: d– singular values, U– left singular vectors, V– right singular vectors
Author(s)
John Fox and Georges Monette
See Also
svd, the standard svd function
Examples
C <- matrix(c(1,2,3,2,5,6,3,6,10), 3, 3) # nonsingular, symmetric
C
SVD(C)
# least squares by the SVD
data("workers")
X <- cbind(1, as.matrix(workers[, c("Experience", "Skill")]))
head(X)
y <- workers$Income
head(y)
(svd <- SVD(X))
VdU <- svd$V %*% diag(1/svd$d) %*%t(svd$U)
(b <- VdU %*% y)
coef(lm(Income ~ Experience + Skill, data=workers))
Solve and Display Solutions for Systems of Linear Simultaneous Equations
Description
Solve the equation system Ax = b, given the coefficient matrix
A and right-hand side vector b, using gaussianElimination.
Display the solutions using showEqn.
Usage
Solve(
A,
b = rep(0, nrow(A)),
vars,
verbose = FALSE,
simplify = TRUE,
fractions = FALSE,
...
)
Arguments
A |
the matrix of coefficients of a system of linear equations |
b |
the vector of constants on the right hand side of the equations. The default is a vector of zeros,
giving the homogeneous equations |
vars |
a numeric or character vector of names of the variables.
If supplied, the length must be equal to the number of unknowns in the equations.
The default is |
verbose |
logical; show the steps of the Gaussian elimination algorithm? |
simplify |
logical; try to simplify the equations? |
fractions |
logical; express numbers as rational fractions, using the |
... |
arguments to be passed to |
Details
This function mimics the base function solve when supplied with two arguments,
(A, b), but gives a prettier result, as a set of equations for the solution. The call
solve(A) with a single argument overloads this, returning the inverse of the matrix A.
For that sense, use the function inv instead.
Value
the function is used primarily for its side effect of printing the solution in a readable form, but it invisibly returns the solution as a character vector
Author(s)
John Fox
See Also
gaussianElimination, showEqn inv, solve
Examples
A1 <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b1 <- c(8, -11, -3)
Solve(A1, b1) # unique solution
A2 <- matrix(1:9, 3, 3)
b2 <- 1:3
Solve(A2, b2, fractions=TRUE) # underdetermined
b3 <- c(1, 2, 4)
Solve(A2, b3, fractions=TRUE) # overdetermined
Calculate the Adjoint of a matrix
Description
This function calculates the adjoint of a square matrix, defined as the transposed matrix of cofactors of all elements.
Usage
adjoint(A)
Arguments
A |
a square matrix |
Value
a matrix of the same size as A
Author(s)
Michael Friendly
See Also
Other determinants:
Det(),
cofactor(),
minor(),
rowCofactors(),
rowMinors()
Examples
A <- J(3, 3) + 2*diag(3)
adjoint(A)
Angle between two vectors
Description
angle calculates the angle between two vectors.
Usage
angle(x, y, degree = TRUE)
Arguments
x |
a numeric vector |
y |
a numeric vector |
degree |
logical; should the angle be computed in degrees?
If |
Value
a scalar containing the angle between the vectors
See Also
Examples
x <- c(2,1)
y <- c(1,1)
angle(x, y) # degrees
angle(x, y, degree = FALSE) # radians
# visually
xlim <- c(0,2.5)
ylim <- c(0,2)
# proper geometry requires asp=1
plot( xlim, ylim, type="n", xlab="X", ylab="Y", asp=1,
main = expression(theta == 18.4))
abline(v=0, h=0, col="gray")
vectors(rbind(x,y), col=c("red", "blue"), cex.lab=c(2, 2))
text(.5, .37, expression(theta))
####
x <- c(-2,1)
y <- c(1,1)
angle(x, y) # degrees
angle(x, y, degree = FALSE) # radians
# visually
xlim <- c(-2,1.5)
ylim <- c(0,2)
# proper geometry requires asp=1
plot( xlim, ylim, type="n", xlab="X", ylab="Y", asp=1,
main = expression(theta == 108.4))
abline(v=0, h=0, col="gray")
vectors(rbind(x,y), col=c("red", "blue"), cex.lab=c(2, 2))
text(0, .4, expression(theta), cex=1.5)
Draw an arc showing the angle between vectors
Description
A utility function for drawing vector diagrams. Draws a circular arc to show the angle between two vectors in 2D or 3D.
Usage
arc(p1, p2, p3, d = 0.1, absolute = TRUE, ...)
Arguments
p1 |
Starting point of first vector |
p2 |
End point of first vector, and also start of second vector |
p3 |
End point of second vector |
d |
The distance from |
absolute |
logical; if |
... |
Details
In this implementation, the two vectors are specified by three points, p1, p2, p3, meaning
a line from p1 to p2, and another line from p2 to p3.
Value
none
References
https://math.stackexchange.com/questions/1507248/find-arc-between-two-tips-of-vectors-in-3d
See Also
Other vector diagrams:
Proj(),
arrows3d(),
circle3d(),
corner(),
plot.regvec3d(),
pointOnLine(),
regvec3d(),
vectors(),
vectors3d()
Examples
library(rgl)
vec <- rbind(diag(3), c(1,1,1))
rownames(vec) <- c("X", "Y", "Z", "J")
open3d()
aspect3d("iso")
vectors3d(vec, col=c(rep("black",3), "red"), lwd=2)
# draw the XZ plane, whose equation is Y=0
planes3d(0, 0, 1, 0, col="gray", alpha=0.2)
# show projections of the unit vector J
segments3d(rbind( c(1,1,1), c(1, 1, 0)))
segments3d(rbind( c(0,0,0), c(1, 1, 0)))
segments3d(rbind( c(1,0,0), c(1, 1, 0)))
segments3d(rbind( c(0,1,0), c(1, 1, 0)))
segments3d(rbind( c(1,1,1), c(1, 0, 0)))
# show some orthogonal vectors
p1 <- c(0,0,0)
p2 <- c(1,1,0)
p3 <- c(1,1,1)
p4 <- c(1,0,0)
# show some angles
arc(p1, p2, p3, d=.2)
arc(p4, p1, p2, d=.2)
arc(p3, p1, p2, d=.2)
Draw 3D arrows
Description
Draws nice 3D arrows with cone3ds at their tips.
Usage
arrows3d(
coords,
headlength = 0.035,
head = "end",
scale = NULL,
radius = NULL,
ref.length = NULL,
draw = TRUE,
...
)
Arguments
coords |
A 2n x 3 matrix giving the start and end (x,y,z) coordinates of n arrows, in pairs. The first vector in each pair is taken as the starting coordinates of the arrow, the second as the end coordinates. |
headlength |
Length of the arrow heads, in device units |
head |
Position of the arrow head. Only |
scale |
Scale factor for base and tip of arrow head, a vector of length 3, giving relative scale factors for X, Y, Z |
radius |
radius of the base of the arrow head |
ref.length |
length of vector to be used to scale all of the arrow heads (permits drawing arrow heads of the same size as in a previous call);
if |
draw |
if |
... |
rgl arguments passed down to |
Details
This function is meant to be analogous to arrows, but for 3D plots using rgl.
headlength, scale and radius set the length, scale factor and base radius of the arrow head, a
3D cone. The units of these are all in terms of the ranges of the current rgl 3D scene.
Value
invisibly returns the length of the vector used to scale the arrow heads
Author(s)
January Weiner, borrowed from the pca3d package, slightly modified by John Fox
See Also
Other vector diagrams:
Proj(),
arc(),
circle3d(),
corner(),
plot.regvec3d(),
pointOnLine(),
regvec3d(),
vectors(),
vectors3d()
Examples
#none yet
Build/Get transformation matrices
Description
Recover the history of the row operations that have been performed. This function combines the transformation matrices into a single transformation matrix representing all row operations or may optionally print all the individual operations which have been performed.
Usage
buildTmat(x, all = FALSE)
## S3 method for class 'trace'
as.matrix(x, ...)
## S3 method for class 'trace'
print(x, ...)
Arguments
x |
a matrix A, joined with a vector of constants, b, that has been passed to
|
all |
logical; print individual transformation ies? |
... |
additional arguments |
Value
the transformation matrix or a list of individual transformation matrices
Author(s)
Phil Chalmers
See Also
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b <- c(8, -11, -3)
# using row operations to reduce below diagonal to 0
Abt <- Ab <- cbind(A, b)
Abt <- rowadd(Abt, 1, 2, 3/2)
Abt <- rowadd(Abt, 1, 3, 1)
Abt <- rowadd(Abt, 2, 3, -4)
Abt
# build T matrix and multiply by original form
(T <- buildTmat(Abt))
T %*% Ab # same as Abt
# print all transformation matrices
buildTmat(Abt, TRUE)
# invert transformation matrix to reverse operations
inv(T) %*% Abt
# gaussian elimination
(soln <- gaussianElimination(A, b))
T <- buildTmat(soln)
inv(T) %*% soln
Cholesky Square Root of a Matrix
Description
Returns the Cholesky square root of the non-singular, symmetric matrix X.
The purpose is mainly to demonstrate the algorithm used by Kennedy & Gentle (1980).
Usage
cholesky(X, tol = sqrt(.Machine$double.eps))
Arguments
X |
a square symmetric matrix |
tol |
tolerance for checking for 0 pivot |
Value
the Cholesky square root of X
Author(s)
John Fox
References
Kennedy W.J. Jr, Gentle J.E. (1980). Statistical Computing. Marcel Dekker.
See Also
chol for the base R function
gsorth for Gram-Schmidt orthogonalization of a data matrix
Examples
C <- matrix(c(1,2,3,2,5,6,3,6,10), 3, 3) # nonsingular, symmetric
C
cholesky(C)
cholesky(C) %*% t(cholesky(C)) # check
Draw circles on an existing plot.
Description
Draw circles on an existing plot.
Usage
circle(
x,
y,
radius,
nv = 60,
border = NULL,
col = NA,
lty = 1,
density = NULL,
angle = 45,
lwd = 1
)
Arguments
x, y |
Coordinates of the center of the circle. If |
radius |
Radius (or radii) of the circle(s) in user units. |
nv |
Number of vertices to draw the circle. |
border |
Color to use for drawing the circumference. |
col |
Color to use for filling the circle. |
lty |
Line type for the circumference. |
density |
Density for patterned fill. See |
angle |
Angle of patterned fill. See |
lwd |
Line width for the circumference. |
Details
Rather than depending on the aspect ratio par("asp") set globally or
in the call to plot,
circle uses the dimensions of the current plot and the x and y coordinates to draw a circle rather than an ellipse.
Of course, if you resize the plot the aspect ratio can change.
This function was copied from draw.circle
Value
Invisibly returns a list with the x and y coordinates of the points on the circumference of the last circle displayed.
Author(s)
Jim Lemon, thanks to David Winsemius for the density and angle args
See Also
Examples
plot(1:5,seq(1,10,length=5),
type="n",xlab="",ylab="",
main="Test draw.circle")
# draw three concentric circles
circle(2, 4, c(1, 0.66, 0.33),border="purple",
col=c("#ff00ff","#ff77ff","#ffccff"),lty=1,lwd=1)
# draw some others
circle(2.5, 8, 0.6,border="red",lty=3,lwd=3)
circle(4, 3, 0.7,border="green",col="yellow",lty=1,
density=5,angle=30,lwd=10)
circle(3.5, 8, 0.8,border="blue",lty=2,lwd=2)
Draw a horizontal circle
Description
A utility function for drawing a horizontal circle in the (x,y) plane in a 3D graph
Usage
circle3d(center, radius, segments = 100, fill = FALSE, ...)
Arguments
center |
A vector of length 3. |
radius |
A positive number. |
segments |
An integer specifying the number of line segments to use to draw the circle (default, 100). |
fill |
logical; if |
... |
rgl material properties for the circle. |
See Also
Other vector diagrams:
Proj(),
arc(),
arrows3d(),
corner(),
plot.regvec3d(),
pointOnLine(),
regvec3d(),
vectors(),
vectors3d()
Examples
ctr=c(0,0,0)
circle3d(ctr, 3, fill = TRUE)
circle3d(ctr - c(-1,-1,0), 3, col="blue")
circle3d(ctr + c(1,1,0), 3, col="red")
Class Data Set
Description
A small artificial data set used to illustrate statistical concepts.
Usage
data("class")Format
A data frame with 15 observations on the following 4 variables.
sexa factor with levels
FMagea numeric vector
heighta numeric vector
weighta numeric vector
Examples
data(class)
plot(class)
Cofactor of A[i,j]
Description
Returns the cofactor of element (i,j) of the square matrix A, i.e., the signed minor of the sub-matrix that results when row i and column j are deleted.
Usage
cofactor(A, i, j)
Arguments
A |
a square matrix |
i |
row index |
j |
column index |
Value
the cofactor of A[i,j]
Author(s)
Michael Friendly
See Also
rowCofactors for all cofactors of a given row
Other determinants:
Det(),
adjoint(),
minor(),
rowCofactors(),
rowMinors()
Examples
M <- matrix(c(4, -12, -4,
2, 1, 3,
-1, -3, 2), 3, 3, byrow=TRUE)
cofactor(M, 1, 1)
cofactor(M, 1, 2)
cofactor(M, 1, 3)
Data on Coffee, Stress and Heart Damage
Description
A dataset, used for examples in Friendly, Fox & Monette (2013),
giving an index of cardiac damage (Heart)
in relation to a measure of daily coffee consumption (Coffee)
and Stress, a measure of perceived occupational stress,
in a contrived sample of n = 20 university people.
Usage
data("coffee")Format
A data frame with 20 observations on the following 4 variables.
Groupuniversity group, a factor with levels
Grad_StudentProfessorStudentCoffeea measure of daily coffee consumption
Stressa measure of perceived occupational stress
Heartan index of cardiac damage
Details
The main goal for analysis of this teaching example would be to determine whether or not coffee is good or bad for your heart, and stress represents one potential confounding variable among others (age, smoking, etc.) that might be useful to control statistically.
Friendly et al. (2013) use this data to illustrate
(a) data ellipses in data space and the corresponding confidence ellipses in
parameter (\beta) space; (b) effects of measurement error in a predictor or response;
(c) added-variable plots and more.
Source
This dataset was constructed by Georges Monette, and was modified from that in his spida2 package,
https://github.com/gmonette/spida2.
References
Friendly, M., Monette, G., & Fox, J. (2013). Elliptical Insights: Understanding Statistical Methods Through Elliptical Geometry. Statistical Science, 28(1), 1–39. https://doi.org/10.1214/12-STS402
Examples
data(coffee)
## maybe str(coffee) ; plot(coffee) ...
Draw a 3D cone
Description
Draws a cone in 3D from a base point to a tip point, with a given radius at the base.
This is used to draw nice arrow heads in arrows3d.
Usage
cone3d(base, tip, radius = 10, col = "grey", scale = NULL, ...)
Arguments
base |
coordinates of base of the cone |
tip |
coordinates of tip of the cone |
radius |
radius of the base |
col |
color |
scale |
scale factor for base and tip |
... |
rgl arguments passed down; see |
Value
returns the integer object ID of the shape that was added to the scene
Author(s)
January Weiner, borrowed from from the pca3d package
See Also
Examples
# none yet
Draw a corner showing the angle between two vectors
Description
A utility function for drawing vector diagrams. Draws two line segments to indicate the angle between two vectors, typically used for indicating orthogonal vectors are at right angles in 2D and 3D diagrams.
Usage
corner(p1, p2, p3, d = 0.1, absolute = TRUE, ...)
Arguments
p1 |
Starting point of first vector |
p2 |
End point of first vector, and also start of second vector |
p3 |
End point of second vector |
d |
The distance from |
absolute |
logical; if |
... |
Details
In this implementation, the two vectors are specified by three points, p1, p2, p3, meaning
a line from p1 to p2, and another line from p2 to p3.
Value
none
See Also
Other vector diagrams:
Proj(),
arc(),
arrows3d(),
circle3d(),
plot.regvec3d(),
pointOnLine(),
regvec3d(),
vectors(),
vectors3d()
Examples
# none yet
Echelon Form of a Matrix
Description
Returns the (reduced) row-echelon form of the matrix A, using gaussianElimination.
Usage
echelon(A, B, reduced = TRUE, ...)
Arguments
A |
coefficient matrix |
B |
right-hand side vector or matrix. If |
reduced |
logical; should reduced row echelon form be returned? If |
... |
other arguments passed to |
Details
When the matrix A is square and non-singular, the reduced row-echelon result will be the
identity matrix, while the row-echelon from will be an upper triangle matrix.
Otherwise, the result will have some all-zero rows, and the rank of the matrix
is the number of not all-zero rows.
Value
the reduced echelon form of X.
Author(s)
John Fox
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b <- c(8, -11, -3)
echelon(A, b, verbose=TRUE, fractions=TRUE) # reduced row-echelon form
echelon(A, b, reduced=FALSE, verbose=TRUE, fractions=TRUE) # row-echelon form
A <- matrix(c(1,2,3,4,5,6,7,8,10), 3, 3) # a nonsingular matrix
A
echelon(A, reduced=FALSE) # the row-echelon form of A
echelon(A) # the reduced row-echelon form of A
b <- 1:3
echelon(A, b) # solving the matrix equation Ax = b
echelon(A, diag(3)) # inverting A
B <- matrix(1:9, 3, 3) # a singular matrix
B
echelon(B)
echelon(B, reduced=FALSE)
echelon(B, b)
echelon(B, diag(3))
Gaussian Elimination
Description
gaussianElimination demonstrates the algorithm of row reduction used for solving
systems of linear equations of the form A x = B. Optional arguments verbose
and fractions may be used to see how the algorithm works.
Usage
gaussianElimination(
A,
B,
tol = sqrt(.Machine$double.eps),
verbose = FALSE,
latex = FALSE,
fractions = FALSE
)
## S3 method for class 'enhancedMatrix'
print(x, ...)
Arguments
A |
coefficient matrix |
B |
right-hand side vector or matrix. If |
tol |
tolerance for checking for 0 pivot |
verbose |
logical; if |
latex |
logical; if |
fractions |
logical; if |
x |
matrix to print |
... |
arguments to pass down |
Value
If B is absent, returns the reduced row-echelon form of A.
If B is present, returns the reduced row-echelon form of A, with the
same operations applied to B.
Author(s)
John Fox
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b <- c(8, -11, -3)
gaussianElimination(A, b)
gaussianElimination(A, b, verbose=TRUE, fractions=TRUE)
gaussianElimination(A, b, verbose=TRUE, fractions=TRUE, latex=TRUE)
# determine whether matrix is solvable
gaussianElimination(A, numeric(3))
# find inverse matrix by elimination: A = I -> A^-1 A = A^-1 I -> I = A^-1
gaussianElimination(A, diag(3))
inv(A)
# works for 1-row systems (issue # 30)
A2 <- matrix(c(1, 1), nrow=1)
b2 = 2
gaussianElimination(A2, b2)
showEqn(A2, b2)
# plotEqn works for this case
plotEqn(A2, b2)
Correct for aspect and coordinate ratio
Description
Calculate a multiplication factor for the Y dimension to correct for unequal plot aspect and coordinate ratios on the current graphics device.
Usage
getYmult()
Details
getYmult retrieves the plot aspect ratio and the coordinate ratio for the current graphics device, calculates a
multiplicative factor to equalize the X and Y dimensions of a plotted graphic object.
Value
The correction factor for the Y dimension.
Author(s)
Jim Lemon
Gram-Schmidt Orthogonalization of a Matrix
Description
Calculates a matrix with uncorrelated columns using the Gram-Schmidt process
Usage
gsorth(y, order, recenter = TRUE, rescale = TRUE, adjnames = TRUE)
Arguments
y |
a numeric matrix or data frame |
order |
if specified, a permutation of the column indices of |
recenter |
logical; if |
rescale |
logical; if |
adjnames |
logical; if |
Details
This function, originally from the heplots package has now been deprecated in matlib. Use
GramSchmidt instead.
Value
a matrix/data frame with uncorrelated columns
Examples
## Not run:
set.seed(1234)
A <- matrix(c(1:60 + rnorm(60)), 20, 3)
cor(A)
G <- gsorth(A)
zapsmall(cor(G))
## End(Not run)
Test for square matrix
Description
Test for square matrix
Test for square, symmetric matrix
Test for orthogonal matrix
Usage
is_square_matrix(x)
is_symmetric_matrix(x)
is_orthogonal_matrix(x)
Arguments
x |
A numeric matrix |
Value
TRUE if x is a square matrix, else FALSE
TRUE if x is a square, symmetric matrix, else FALSE
TRUE if x is an orthogonal matrix, else FALSE
Create and Manipulate LaTeX Representations of Matrices
Description
The purpose of the latexMatrix() function is to facilitate the preparation
of LaTeX and Markdown documents that include matrices. The function generates the
the LaTeX code for matrices of various types programmatically. The objects produced
by the function can also be manipulated, e.g., with standard arithmetic functions and operators:
See latexMatrixOperations.
The latexMatrix() function can construct the LaTeX code for a symbolic matrix
whose elements are a symbol, like x, with row and column subscripts.
For example, with no arguments, the call latexMatrix() generates this LaTeX representation
of an n \times m matrix with elements x_{ij}.
\begin{pmatrix}
x_{11} & x_{12} & \dots & x_{1m} \
x_{21} & x_{22} & \dots & x_{2m} \
\vdots & \vdots & \ddots & \vdots \
x_{n1} & x_{n2} & \dots & x_{nm}
\end{pmatrix}
When rendered in LaTeX, this produces:
\begin{pmatrix}
x_{11} & x_{12} & \cdots & x_{1m} \\
x_{21} & x_{22} & \cdots & x_{2m} \\
\vdots & \vdots & & \vdots \\
x_{n1} & x_{n2} & \cdots & x_{nm} \\
\end{pmatrix}
Alternatively, instead of characters,
the number of rows and/or columns can be integers, generating a matrix of given size,
as in latexMatrix(nrow = 2, ncol = 3).
As well, instead of a character for the matrix symbol, you can supply a matrix of arbitrary character
strings (in LaTeX notation) or numbers, and these will be used as the elements of the matrix,
as in latexMatrix(matrix(1:6, nrow = 2, ncol = 6)).
The resulting LaTeX code is printed to the console by default. When the result is assigned to a variable,
you can send it to the clipboard using write_clip(). Perhaps most convenient of all,
the function can be used used in a markdown chunk in a Rmd or qmd document, e.g,
```{r results = "asis"}
latexMatrix("\\lambda", nrow=2, ncol=2,
diag=TRUE)
```
This generates
\begin{pmatrix}
\lambda_{1} & 0 \\
0 & \lambda_{2} \\
\end{pmatrix}
The function Eqn can be used to construct matrix equations, and in RStudio generates a preview of
an equation in the Viewer panel.
Various options control the printing of "latexMatrix" objects, described in Details.
Usage
latexMatrix(
symbol = "x",
nrow = "n",
ncol = "m",
rownames = NULL,
colnames = NULL,
matrix = getOption("latexMatrixEnv"),
diag = FALSE,
sparse = FALSE,
zero.based = c(FALSE, FALSE),
end.at = c("n - 1", "m - 1"),
comma = any(zero.based),
exponent,
transpose = FALSE,
show.size = FALSE,
digits = getOption("digits") - 2,
fractions = FALSE,
prefix = "",
suffix = "",
prefix.row = "",
prefix.col = ""
)
partition(x, ...)
## S3 method for class 'latexMatrix'
partition(x, rows, columns, ...)
getLatex(x, ...)
## S3 method for class 'latexMatrix'
getLatex(x, ...)
getBody(x, ...)
## S3 method for class 'latexMatrix'
getBody(x, ...)
getWrapper(x, ...)
## S3 method for class 'latexMatrix'
getWrapper(x, ...)
Dim(x, ...)
## S3 method for class 'latexMatrix'
Dim(x, ...)
Nrow(x, ...)
## S3 method for class 'latexMatrix'
Nrow(x, ...)
Ncol(x, ...)
## S3 method for class 'latexMatrix'
Ncol(x, ...)
## S3 method for class 'latexMatrix'
print(
x,
onConsole = TRUE,
bordermatrix = getOption("print.latexMatrix")[["bordermatrix"]],
cell.spacing = getOption("print.latexMatrix")[["cell.spacing"]],
colname.spacing = getOption("print.latexMatrix")[["colname.spacing"]],
text.labels = getOption("print.latexMatrix")[["text.labels"]],
display.labels = getOption("print.latexMatrix")[["display.labels"]],
mathtext = getOption("print.latexMatrix")[["mathtext"]],
mathtext.size = getOption("print.latexMatrix")[["mathtext.size"]],
...
)
## S3 method for class 'latexMatrix'
is.numeric(x)
## S3 method for class 'latexMatrix'
as.double(x, locals = list(), ...)
## S3 method for class 'latexMatrix'
x[i, j, ..., drop]
## S3 method for class 'latexMatrix'
cbind(..., deparse.level)
## S3 method for class 'latexMatrix'
rbind(..., deparse.level)
## S3 method for class 'latexMatrix'
dimnames(x)
Dimnames(x) <- value
## S3 replacement method for class 'latexMatrix'
Dimnames(x) <- value
Rownames(x) <- value
## S3 replacement method for class 'latexMatrix'
Rownames(x) <- value
Colnames(x) <- value
## S3 replacement method for class 'latexMatrix'
Colnames(x) <- value
Arguments
symbol |
name for matrix elements, character string. For LaTeX symbols,
the backslash must be doubled because it is an escape character in R.
That is, you must use |
nrow |
Number of rows, a single character representing rows symbolically, or an integer, generating that many rows. |
ncol |
Number of columns, a single character representing columns symbolically, or an integer, generating that many columns. |
rownames |
optional vector of names for the matrix rows.
if |
colnames |
optional vector of names for the matrix columns.
if |
matrix |
Character string giving the LaTeX matrix environment used in
Small matrix definitions from the |
diag |
logical; if |
sparse |
logical; if |
zero.based |
logical 2-vector; start the row and/or column indices at 0 rather than 1;
the default is |
end.at |
if row or column indices start at 0, should they end at |
comma |
logical; if |
exponent |
if specified, e.g., |
transpose |
if |
show.size |
logical; if |
digits |
for a numeric matrix, number of digits to display; |
fractions |
logical; if |
prefix |
optional character string to be pre-pended to each matrix element, e.g, to wrap each
element in a function like |
suffix |
optional character string to be appended to each matrix element, e.g., for exponents on each element |
prefix.row |
optional character string to be pre-pended to each matrix row index |
prefix.col |
optional character string to be pre-pended to each matrix column index |
x |
a |
... |
for |
rows |
row numbers after which partition lines should be drawn in the LaTeX printed representation of the matrix; if omitted, then the matrix isn't partitioned by rows |
columns |
column numbers after which partition lines should be drawn in the LaTeX printed representation of the matrix; if omitted, then the matrix isn't partitioned by columns |
onConsole |
if |
bordermatrix |
if |
cell.spacing |
a character whose width is used to try to even out spacing
of printed cell elements; the default is taken from the |
colname.spacing |
a character whose width is used to try to even out spacing
of printed column names; the default is taken from the |
text.labels |
whether to set row and column labels in text mode rather than
math model; the default is taken from the |
display.labels |
whether or not to display row and column labels (if they exist);
the default is taken from the |
mathtext |
a LaTeX command to display row/column label text in math mode;
the default is taken from the |
mathtext.size |
a LaTeX command to control the size of row/column text
(e.g., |
locals |
an optional list or named numeric vector of variables to be given
specific numeric values; e.g.,
|
i |
row index or indices (negative indices to omit rows) |
j |
column index or indices (negative indices to omit columns) |
drop |
to match the generic indexing function, ignored |
deparse.level |
|
value |
for |
Details
This implementation assumes that the LaTeX amsmath package will be available because it uses the shorthands
\begin{pmatrix}, ... rather than
\left(
\begin{array}(ccc)
...
\end{array}
\right)
You may need to use extra_dependencies: ["amsmath"] in your YAML header of a Rmd or qmd file.
You can supply a numeric matrix as the symbol, but the result will not be pretty
unless the elements are integers or are rounded. You can control the number of digits
displayed using the global option options("digits"), for example: options(digits = 4).
For a LaTeX representation of general numeric matrices, use
matrix2latex.
Other functions
rbind() and cbind() join "latexMatrix" objects together, and indexing,
via [ , ] subsets rows/columns just as they do for regular matrices.
The partition() function modifies (only) the printed LaTeX representation of a "latexMatrix"
object to include partition lines by rows and/or columns.
The accessor functions getLatex(), getBody(), getWrapper(),
getDim(), getNrow(), and getNcol() may be used to retrieve
components of the returned object.
Various arithmetic functions and operators (like +, -, matrix product %*%, ...) for "latexMatrix" objects are
documented separately; see, latexMatrixOperations.
print.latexMatrix options
Some LaTeX typesetting details are controlled by the "print.latexMatrix" option,
which can be a list with one or more of the following elements (see the
arguments to the print.latexMatrix() method for more information):
"bordermatrix",
"cell.spacing",
"colname.spacing",
"text.labels",
"display.labels",
"mathtext",
and "mathtext.size".
Most of these have to do with the display of matrices which have row and/or column labels
in their dimnames or by being set with the rownames and rownames
arguments to latexMatrix.
You can turn off their display using:
options(print.latexMatrix = list(display.labels=FALSE))
and similarly you can change any other of these options.
Value
latexMatrix() returns an object of class "latexMatrix".
This is a list which contains the LaTeX representation of the matrix as a character string
and other information.
The elements in the returned object are named:
-
"matrix"(the LaTeX representation of the matrix); -
"dim"(nrowandncol); -
"body"(a character matrix of LaTeX expressions for the cells of the matrix); -
"wrapper"(the beginning and ending lines for the LaTeX matrix environment). -
"dimnames"(the rownames and colnames for the matrix, if specified)
partition(), rbind(), cbind(), and indexing of
"latexMatrix" objects also return a "latexMatrix" object.
Author(s)
John Fox
See Also
latexMatrixOperations,
Eqn,
matrix2latex,
write_clip
Examples
latexMatrix()
# return value
mat <- latexMatrix()
str(mat)
cat(getLatex(mat))
# copy to clipboard (can't be done in non-interactive mode)
## Not run:
clipr::write_clip(mat)
## End(Not run)
# can use a complex symbol
latexMatrix("\\widehat{\\beta}", 2, 4)
# numeric rows/cols
latexMatrix(ncol=3)
latexMatrix(nrow=4)
latexMatrix(nrow=4, ncol=4)
# diagonal matrices
latexMatrix(nrow=3, ncol=3, diag=TRUE)
latexMatrix(nrow="n", ncol="n", diag=TRUE)
latexMatrix(nrow="n", ncol="n", diag=TRUE, sparse=TRUE)
# commas, exponents, transpose
latexMatrix("\\beta", comma=TRUE, exponent="-1")
latexMatrix("\\beta", comma=TRUE, transpose=TRUE)
latexMatrix("\\beta", comma=TRUE, exponent="-1", transpose=TRUE)
# for a row/column vector, wrap in matrix()
latexMatrix(matrix(LETTERS[1:4], nrow=1))
latexMatrix(matrix(LETTERS[1:4], ncol=1))
# represent the SVD, X = U D V' symbolically
X <- latexMatrix("x", "n", "p")
U <- latexMatrix("u", "n", "k")
D <- latexMatrix("\\lambda", "k", "k", diag=TRUE)
V <- latexMatrix("v", "k", "p", transpose = TRUE)
cat("\\mathrm{SVD:}\n", getLatex(X), "=\n", getLatex(U),
getLatex(D), getLatex(V))
# supply a matrix for 'symbol'
m <- matrix(c(
"\\alpha", "\\beta",
"\\gamma", "\\delta",
"\\epsilon", "\\pi",
0 , 0), 4, 2, byrow=TRUE)
latexMatrix(m)
# Identity matrix
latexMatrix(diag(3))
latexMatrix(diag(3), sparse=TRUE)
# prefix / suffix
latexMatrix(prefix="\\sqrt{", suffix="}")
latexMatrix(suffix="^{1/2}")
# show size (order) of a matrix
latexMatrix(show.size=TRUE)
latexMatrix(nrow=3, ncol=4, show.size=TRUE)
# handling fractions
m <- matrix(3/(1:9), 3, 3)
latexMatrix(m)
latexMatrix(m, digits=2)
latexMatrix(m, fractions=TRUE)
# zero-based indexing
latexMatrix(zero.based=c(TRUE, TRUE))
# partitioned matrix
X <- latexMatrix(nrow=5, ncol=6)
partition(X, rows=c(2, 4), columns=c(3, 5))
# binding rows and columns; indexing
X <- latexMatrix("x", nrow=4, ncol=2)
Y <- latexMatrix("y", nrow=4, ncol=1)
Z <- latexMatrix(matrix(1:8, 4, 2))
cbind(X, Y, Z)
rbind(X, Z)
X[1:2, ]
X[-(1:2), ]
X[1:2, 2]
# defining row and column names
W <- latexMatrix(rownames=c("\\alpha_1", "\\alpha_2", "\\alpha_m"),
colnames=c("\\beta_1", "\\beta_2", "\\beta_n"))
W
Rownames(W) <- c("\\mathrm{Abe}", "\\mathrm{Barry}", "\\mathrm{Zelda}")
Colnames(W) <- c("\\mathrm{Age}", "\\mathrm{BMI}", "\\mathrm{Waist}")
W
Various Functions and Operators for "latexMatrix" Objects
Description
These operators and functions provide for LaTeX representations of symbolic and numeric matrix arithmetic and computations. They provide reasonable means to compose meaningful matrix equations in LaTeX far easier than doing this manually matrix by matrix.
The following operators and functions are documented here:
-
matsum()and+, matrix addition; -
matdiff()and-, matrix subtraction and negation; -
*, product of a scalar and a matrix; -
Dot(), inner product of two vectors; -
matprod()and%*%, matrix product; -
matpower()and^, powers (including inverse) of a square matrix; -
solve()andinverse(), matrix inverse of a square matrix; -
t(), transpose; -
determinant()of a square matrix; -
kronecker()and%O%, the Kronecker product.
Usage
matsum(A, ...)
## S3 method for class 'latexMatrix'
matsum(A, ..., as.numeric = TRUE)
## S3 method for class 'latexMatrix'
e1 + e2
matdiff(A, B, ...)
## S3 method for class 'latexMatrix'
matdiff(A, B = NULL, as.numeric = TRUE, ...)
## S3 method for class 'latexMatrix'
e1 - e2
## S3 method for class 'latexMatrix'
e1 * e2
Dot(x, y, simplify = TRUE)
matmult(X, ...)
## S3 method for class 'latexMatrix'
matmult(X, ..., simplify = TRUE, as.numeric = TRUE)
## S3 method for class 'latexMatrix'
x %*% y
matpower(X, power, ...)
## S3 method for class 'latexMatrix'
matpower(X, power, simplify = TRUE, as.numeric = TRUE, ...)
## S3 method for class 'latexMatrix'
e1 ^ e2
inverse(X, ...)
## S3 method for class 'latexMatrix'
inverse(X, ..., as.numeric = TRUE, simplify = TRUE)
## S3 method for class 'latexMatrix'
t(x)
## S3 method for class 'latexMatrix'
determinant(x, logarithm, ...)
## S3 method for class 'latexMatrix'
solve(
a,
b,
simplify = FALSE,
as.numeric = TRUE,
frac = c("\\dfrac", "\\frac", "\\tfrac", "\\cfrac"),
...
)
## S4 method for signature 'latexMatrix,latexMatrix'
kronecker(X, Y, FUN = "*", make.dimnames = FALSE, ...)
x %X% y
Arguments
A |
a |
... |
for |
as.numeric |
if |
e1 |
a |
e2 |
a |
B |
a |
x |
for |
y |
for |
simplify |
if |
X |
a |
power |
to raise a square matrix to this power, an integer |
logarithm |
to match the generic |
a |
a |
b |
ignored; to match the |
frac |
LaTeX command to use in forming fractions; the default
is |
Y |
a |
FUN |
to match the |
make.dimnames |
to match the |
Details
These operators and functions only apply to "latexMatrix" objects
of definite (i.e., numeric) dimensions.
When there are both a function and an
operator (e.g., matmult() and %*%), the former is more
flexible via optional arguments and the latter calls the former with default
arguments. For example, using the operator A %*% B multiplies
the two matrices A and B, returning a symbolic result.
The function matmult() multiplies two or more matrices, and
can simplify the result and/or produced the numeric representation of the
product.
The result of matrix multiplication, \mathbf{C} = \mathbf{A} \: \mathbf{B}
is composed of the vector inner (dot) products of each row of \mathbf{A} with
each column of \mathbf{B},
c_{ij} = \mathbf{a}_i^\top \mathbf{b}_j
= \Sigma_k a_{ik} \cdot b_{kj}
The Dot() function computes the inner product symbolically in LaTeX notation for
numeric and character vectors, simplifying the result if simplify = TRUE.
The LaTeX symbol for multiplication ("\cdot" by default)
can be changed by changing options(latexMultSymbol),
e.g, options(latexMultSymbol = "\\times") (note the double-backslash).
Value
All of these functions return "latexMatrix" objects,
except for Dot(), which returns a LaTeX expression as a character string.
Author(s)
John Fox
See Also
Examples
A <- latexMatrix(symbol="a", nrow=2, ncol=2)
B <- latexMatrix(symbol="b", nrow=2, ncol=2)
A
B
A + B
A - B
"a" * A
C <- latexMatrix(symbol="c", nrow=2, ncol=3)
A %*% C
t(C)
determinant(A)
cat(solve(A, simplify=TRUE))
D <- latexMatrix(matrix(letters[1:4], 2, 2))
D
as.numeric(D, locals=list(a=1, b=2, c=3, d=4))
X <- latexMatrix(matrix(c(3, 2, 0, 1, 1, 1, 2,-2, 1), 3, 3))
X
as.numeric(X)
MASS::fractions(as.numeric(inverse(X)))
(d <- determinant(X))
eval(parse(text=(gsub("\\\\cdot", "*", d))))
X <- latexMatrix(matrix(1:6, 2, 3), matrix="bmatrix")
I3 <- latexMatrix(diag(3))
I3 %X% X
kronecker(I3, X, sparse=TRUE)
(E <- latexMatrix(diag(1:3)))
# equivalent:
X %*% E
matmult(X, E)
matmult(X, E, simplify=FALSE, as.numeric=FALSE)
# equivalent:
X %*% E %*% E
matmult(X, E, E)
# equivalent:
E^-1
inverse(E)
solve(E)
solve(E, as.numeric=FALSE) # details
# equivalent
E^3
matpower(E, 3)
matpower(E, 3, as.numeric=FALSE)
Length of a Vector or Column Lengths of a Matrix
Description
len calculates the Euclidean length (also called Euclidean norm) of a vector or the
length of each column of a numeric matrix.
Usage
len(X)
Arguments
X |
a numeric vector or matrix |
Value
a scalar or vector containing the length(s)
See Also
norm for more general matrix norms
Examples
len(1:3)
len(matrix(1:9, 3, 3))
# distance between two vectors
len(1:3 - c(1,1,1))
(Deprecated) Convert matrix to LaTeX equation
Description
(This function has been deprecated; see latexMatrix instead).
This function provides a soft-wrapper to xtable::xtableMatharray() with additional support for
fractions output and brackets.
Usage
matrix2latex(
x,
fractions = FALSE,
brackets = TRUE,
show.size = FALSE,
digits = NULL,
print = TRUE,
...
)
Arguments
x |
a numeric or character matrix. If the latter a numeric-based arguments will be ignored |
fractions |
logical; if |
brackets |
logical or a character in |
show.size |
logical; if |
digits |
Number of digits to display. If |
print |
logical; print the LaTeX code for the matrix on the console?; default: |
... |
additional arguments passed to |
Details
The code for brackets matches some of the options from the AMS matrix LaTeX package:
\pmatrix{}, \bmatrix{}, \Bmatrix{}, ... .
Author(s)
Phil Chalmers
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b <- c(8, -11, -3)
matrix2latex(cbind(A,b))
matrix2latex(cbind(A,b), digits = 0)
matrix2latex(cbind(A/2,b), fractions = TRUE)
matrix2latex(A, digits=0, brackets="p", show.size = TRUE)
# character matrices
A <- matrix(paste0('a_', 1:9), 3, 3)
matrix2latex(cbind(A,b))
b <- paste0("\\beta_", 1:3)
matrix2latex(cbind(A,b))
Minor of A[i,j]
Description
Returns the minor of element (i,j) of the square matrix A, i.e., the determinant of the sub-matrix that results when row i and column j are deleted.
Usage
minor(A, i, j)
Arguments
A |
a square matrix |
i |
row index |
j |
column index |
Value
the minor of A[i,j]
Author(s)
Michael Friendly
See Also
rowMinors for all minors of a given row
Other determinants:
Det(),
adjoint(),
cofactor(),
rowCofactors(),
rowMinors()
Examples
M <- matrix(c(4, -12, -4,
2, 1, 3,
-1, -3, 2), 3, 3, byrow=TRUE)
minor(M, 1, 1)
minor(M, 1, 2)
minor(M, 1, 3)
Matrix Power
Description
A simple function to demonstrate calculating the power of a square symmetric matrix in terms of its eigenvalues and eigenvectors.
Usage
mpower(A, p, tol = sqrt(.Machine$double.eps))
Arguments
A |
a square symmetric matrix |
p |
matrix power, not necessarily a positive integer |
tol |
tolerance for determining if the matrix is symmetric |
Details
The matrix power p can be a fraction or other non-integer. For example, p=1/2 and
p=1/3 give a square-root and cube-root of the matrix.
Negative powers are also allowed. For example, p=-1 gives the inverse and p=-1/2
gives the inverse square-root.
Value
A raised to the power p: A^p
See Also
The {%^%} operator in the expm package is far more efficient
Examples
C <- matrix(c(1,2,3,2,5,6,3,6,10), 3, 3) # nonsingular, symmetric
C
mpower(C, 2)
zapsmall(mpower(C, -1))
solve(C) # check
Plot method for regvec3d objects
Description
The plot method for regvec3d objects uses the low-level graphics tools in this package to draw 3D and 3D
vector diagrams reflecting the partial and marginal
relations of y to x1 and x2 in a bivariate multiple linear regression model,
lm(y ~ x1 + x2).
The summary method prints the vectors and their vector lengths, followed by the summary
for the model.
Usage
## S3 method for class 'regvec3d'
plot(
x,
y,
dimension = 3,
col = c("black", "red", "blue", "brown", "lightgray"),
col.plane = "gray",
cex.lab = 1.2,
show.base = 2,
show.marginal = FALSE,
show.hplane = TRUE,
show.angles = TRUE,
error.sphere = c("none", "e", "y.hat"),
scale.error.sphere = x$scale,
level.error.sphere = 0.95,
grid = FALSE,
add = FALSE,
...
)
## S3 method for class 'regvec3d'
summary(object, ...)
## S3 method for class 'regvec3d'
print(x, ...)
Arguments
x |
A “regvec3d” object |
y |
Ignored; only included for compatibility with the S3 generic |
dimension |
Number of dimensions to plot: |
col |
A vector of 5 colors. |
col.plane |
Color of the base plane in a 3D plot or axes in a 2D plot |
cex.lab |
character expansion applied to vector labels. May be a number or numeric vector corresponding to the the
rows of |
show.base |
If |
show.marginal |
If |
show.hplane |
If |
show.angles |
If |
error.sphere |
Plot a sphere (or in 2D, a circle) of radius proportional to the length of
the residual vector, centered either at the origin ( |
scale.error.sphere |
Whether to scale the error sphere if |
level.error.sphere |
The confidence level for the error sphere, applied if |
grid |
If |
add |
If |
... |
Parameters passed down to functions [unused now] |
object |
A |
Details
A 3D diagram shows the vector y and the plane formed by the predictors,
x1 and x2, where all variables are represented in deviation form, so that
the intercept need not be included.
A 2D diagram, using the first two columns of the result, can be used to show the projection
of the space in the x1, x2 plane.
The drawing functions vectors and vectors3d used by the plot.regvec3d method only work
reasonably well if the variables are shown on commensurate scales, i.e., with
either scale=TRUE or normalize=TRUE.
Value
None
References
Fox, J. (2016). Applied Regression Analysis and Generalized Linear Models, 3rd ed., Sage, Chapter 10.
See Also
Other vector diagrams:
Proj(),
arc(),
arrows3d(),
circle3d(),
corner(),
pointOnLine(),
regvec3d(),
vectors(),
vectors3d()
Examples
if (require(carData)) {
data("Duncan", package="carData")
dunc.reg <- regvec3d(prestige ~ income + education, data=Duncan)
plot(dunc.reg)
plot(dunc.reg, dimension=2)
plot(dunc.reg, error.sphere="e")
summary(dunc.reg)
# Example showing Simpson's paradox
data("States", package="carData")
states.vec <- regvec3d(SATM ~ pay + percent, data=States, scale=TRUE)
plot(states.vec, show.marginal=TRUE)
plot(states.vec, show.marginal=TRUE, dimension=2)
summary(states.vec)
}
Plot Linear Equations
Description
Shows what matrices A, b look like as the system of linear equations, A x = b with two unknowns,
x1, x2, by plotting a line for each equation.
Usage
plotEqn(
A,
b,
vars,
xlim,
ylim,
col = 1:nrow(A),
lwd = 2,
lty = 1,
axes = TRUE,
labels = TRUE,
solution = TRUE,
...
)
Arguments
A |
either the matrix of coefficients of a system of linear equations, or the matrix |
b |
if supplied, the vector of constants on the right hand side of the equations, of length matching
the number of rows of |
vars |
a numeric or character vector of names of the variables.
If supplied, the length must be equal to the number of unknowns in the equations, i.e., 2.
The default is |
xlim |
horizontal axis limits for the first variable |
ylim |
vertical axis limits for the second variable; if missing, |
col |
scalar or vector of colors for the lines, recycled as necessary |
lwd |
scalar or vector of line widths for the lines, recycled as necessary |
lty |
scalar or vector of line types for the lines, recycled as necessary |
axes |
logical; draw horizontal and vertical axes through (0,0)? |
labels |
logical, or a vector of character labels for the equations; if |
solution |
logical; should the solution points for pairs of equations be marked? |
... |
Other arguments passed to |
Value
nothing; used for the side effect of making a plot
Author(s)
Michael Friendly
References
Fox, J. and Friendly, M. (2016). "Visualizing Simultaneous Linear Equations, Geometric Vectors, and Least-Squares Regression with the matlib Package for R". useR Conference, Stanford, CA, June 27 - June 30, 2016.
See Also
showEqn, vignette("linear-equations", package="matlib")
Examples
# consistent equations
A<- matrix(c(1,2,3, -1, 2, 1),3,2)
b <- c(2,1,3)
showEqn(A, b)
plotEqn(A,b)
# inconsistent equations
b <- c(2,1,6)
showEqn(A, b)
plotEqn(A,b)
Plot Linear Equations in 3D
Description
Shows what matrices A, b look like as the system of linear equations, A x = b with three unknowns,
x1, x2, and x3, by plotting a plane for each equation.
Usage
plotEqn3d(
A,
b,
vars,
xlim = c(-2, 2),
ylim = c(-2, 2),
zlim,
col = 2:(nrow(A) + 1),
alpha = 0.9,
labels = FALSE,
solution = TRUE,
axes = TRUE,
lit = FALSE
)
Arguments
A |
either the matrix of coefficients of a system of linear equations, or the matrix |
b |
if supplied, the vector of constants on the right hand side of the equations, of length matching
the number of rows of |
vars |
a numeric or character vector of names of the variables.
If supplied, the length must be equal to the number of unknowns in the equations.
The default is |
xlim |
axis limits for the first variable |
ylim |
axis limits for the second variable |
zlim |
horizontal axis limits for the second variable; if missing, |
col |
scalar or vector of colors for the lines, recycled as necessary |
alpha |
transparency applied to each plane |
labels |
logical, or a vector of character labels for the equations; not yet implemented. |
solution |
logical; should the solution point for all equations be marked (if possible) |
axes |
logical; whether to frame the plot with coordinate axes |
lit |
logical, specifying if lighting calculation should take place on geometry; see |
Value
nothing; used for the side effect of making a plot
Author(s)
Michael Friendly, John Fox
References
Fox, J. and Friendly, M. (2016). "Visualizing Simultaneous Linear Equations, Geometric Vectors, and Least-Squares Regression with the matlib Package for R". useR Conference, Stanford, CA, June 27 - June 30, 2016.
Examples
# three consistent equations in three unknowns
A <- matrix(c(13, -4, 2, -4, 11, -2, 2, -2, 8), 3,3)
b <- c(1,2,4)
plotEqn3d(A,b)
Position of a point along a line
Description
A utility function for drawing vector diagrams. Find position of an interpolated point along a line from x1 to x2.
Usage
pointOnLine(x1, x2, d, absolute = TRUE)
Arguments
x1 |
A vector of length 2 or 3, representing the starting point of a line in 2D or 3D space |
x2 |
A vector of length 2 or 3, representing the ending point of a line in 2D or 3D space |
d |
The distance along the line from |
absolute |
logical; if |
Details
The function takes a step of length d along the line defined by the difference between the two points, x2 - x1.
When absolute=FALSE, this step is proportional to the difference,
while when absolute=TRUE, the difference is first scaled to unit length so that the step is always of length d.
Note that the physical length of a line in different directions in a graph depends on the aspect ratio of the plot axes,
and lines of the same length will only appear equal if the aspect ratio is one
(asp=1 in 2D, or aspect3d("iso") in 3D).
Value
The interpolated point, a vector of the same length as x1
See Also
Other vector diagrams:
Proj(),
arc(),
arrows3d(),
circle3d(),
corner(),
plot.regvec3d(),
regvec3d(),
vectors(),
vectors3d()
Examples
x1 <- c(0, 0)
x2 <- c(1, 4)
pointOnLine(x1, x2, 0.5)
pointOnLine(x1, x2, 0.5, absolute=FALSE)
pointOnLine(x1, x2, 1.1)
y1 <- c(1, 2, 3)
y2 <- c(3, 2, 1)
pointOnLine(y1, y2, 0.5)
pointOnLine(y1, y2, 0.5, absolute=FALSE)
Power Method for Eigenvectors
Description
Finds a dominant eigenvalue, \lambda_1, and its corresponding
eigenvector, v_1, of a square matrix by applying Hotelling's (1933) Power Method with scaling.
Usage
powerMethod(A, v = NULL, eps = 1e-06, maxiter = 100, plot = FALSE)
Arguments
A |
a square numeric matrix |
v |
optional starting vector; if not supplied, it uses a unit vector of length equal to the number of rows / columns of |
eps |
convergence threshold for terminating iterations |
maxiter |
maximum number of iterations |
plot |
logical; if |
Details
The method is based upon the fact that repeated multiplication of a matrix A by a trial
vector v_1^{(k)} converges to the value of the eigenvector,
v_1^{(k+1)} = A v_1^{(k)} / \vert\vert A v_1^{(k)} \vert\vert
The corresponding eigenvalue is then found as
\lambda_1 = \frac{v_1^T A v_1}{v_1^T v_1}
In pre-computer days, this method could be extended to find subsequent eigenvalue - eigenvector
pairs by "deflation", i.e., by applying the method again to the new matrix.
A - \lambda_1 v_1 v_1^{T} .
This method is still used in some computer-intensive applications with huge matrices where only the dominant eigenvector is required, e.g., the Google Page Rank algorithm.
Value
a list containing the eigenvector (vector), eigenvalue (value), iterations (iter),
and iteration history (vector_iterations)
Author(s)
Gaston Sanchez (from matrixkit)
References
Hotelling, H. (1933). Analysis of a complex of statistical variables into principal components. Journal of Educational Psychology, 24, 417-441, and 498-520.
Examples
A <- cbind(c(7, 3), c(3, 6))
powerMethod(A)
eigen(A)$values[1] # check
eigen(A)$vectors[,1]
# demonstrate how the power method converges to a solution
powerMethod(A, v = c(-.5, 1), plot = TRUE)
B <- cbind(c(1, 2, 0), c(2, 1, 3), c(0, 3, 1))
(rv <- powerMethod(B))
# deflate to find 2nd latent vector
l <- rv$value
v <- c(rv$vector)
B1 <- B - l * outer(v, v)
powerMethod(B1)
eigen(B)$vectors # check
# a positive, semi-definite matrix, with eigenvalues 12, 6, 0
C <- matrix(c(7, 4, 1, 4, 4, 4, 1, 4, 7), 3, 3)
eigen(C)$vectors
powerMethod(C)
Print Matrices or Matrix Operations Side by Side
Description
This function is designed to print a collection of matrices, vectors, character strings and matrix expressions side by side. A typical use is to illustrate matrix equations in a compact and comprehensible way.
Usage
printMatEqn(..., space = 1, tol = sqrt(.Machine$double.eps), fractions = FALSE)
Arguments
... |
matrices and character operations to be passed and printed to the console. These
can include named arguments, character string operation symbols (e.g., |
space |
amount of blank spaces to place around operations such as |
tol |
tolerance for rounding |
fractions |
logical; if |
Value
NULL; A formatted sequence of matrices and matrix operations is printed to the console
Author(s)
Phil Chalmers
See Also
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
x <- c(2, 3, -1)
# provide implicit or explicit labels
printMatEqn(AA = A, "*", xx = x, '=', b = A %*% x)
printMatEqn(A, "*", x, '=', b = A %*% x)
printMatEqn(A, "*", x, '=', A %*% x)
# compare with showEqn
b <- c(4, 2, 1)
printMatEqn(A, x=paste0("x", 1:3),"=", b)
showEqn(A, b)
# decimal example
A <- matrix(c(0.5, 1, 3, 0.75, 2.8, 4), nrow = 2)
x <- c(0.5, 3.7, 2.3)
y <- c(0.7, -1.2)
b <- A %*% x - y
printMatEqn(A, "*", x, "-", y, "=", b)
printMatEqn(A, "*", x, "-", y, "=", b, fractions=TRUE)
(Deprecated) Print a matrix, allowing fractions or LaTeX output
Description
(Deprecated) Print a matrix, allowing fractions or LaTeX output
Usage
printMatrix(
A,
parent = TRUE,
fractions = FALSE,
latex = FALSE,
tol = sqrt(.Machine$double.eps)
)
Arguments
A |
A numeric matrix |
parent |
flag used to search in the parent envir for suitable definitions of other arguments.
Set to |
fractions |
If |
latex |
If |
tol |
Tolerance for rounding small numbers to 0 |
Value
The formatted matrix
See Also
Examples
A <- matrix(1:12, 3, 4) / 6
printMatrix(A, fractions=TRUE)
printMatrix(A, latex=TRUE)
Vector space representation of a two-variable regression model
Description
regvec3d calculates the 3D vectors that represent the projection of a two-variable multiple
regression model from n-D observation space into the 3D mean-deviation variable space that they span, thus
showing the regression of y on x1 and x2 in the model lm(y ~ x1 + x2).
The result can be used to draw 2D and 3D vector diagrams accurately reflecting the partial and marginal
relations of y to x1 and x2 as vectors in this representation.
Usage
regvec3d(x1, ...)
## S3 method for class 'formula'
regvec3d(
formula,
data = NULL,
which = 1:2,
name.x1,
name.x2,
name.y,
name.e,
name.y.hat,
name.b1.x1,
name.b2.x2,
abbreviate = 0,
...
)
## Default S3 method:
regvec3d(
x1,
x2,
y,
scale = FALSE,
normalize = TRUE,
name.x1 = deparse(substitute(x1)),
name.x2 = deparse(substitute(x2)),
name.y = deparse(substitute(y)),
name.e = "residuals",
name.y.hat = paste0(name.y, "hat"),
name.b1.x1 = paste0("b1", name.x1),
name.b2.x2 = paste0("b2", name.x2),
name.y1.hat = paste0(name.y, "hat 1"),
name.y2.hat = paste0(name.y, "hat 2"),
...
)
Arguments
x1 |
The generic argument or the first predictor passed to the default method |
... |
Arguments passed to methods |
formula |
A two-sided formula for the linear regression model. It must contain two quantitative predictors
( |
data |
A data frame in which the variables in the model are found |
which |
Indices of predictors variables in the model taken as |
name.x1 |
Name for |
name.x2 |
Ditto for the name of |
name.y |
Ditto for the name of |
name.e |
Name for the residual vector. Default: |
name.y.hat |
Name for the fitted vector |
name.b1.x1 |
Name for the vector corresponding to the partial coefficient of |
name.b2.x2 |
Name for the vector corresponding to the partial coefficient of |
abbreviate |
An integer. If |
x2 |
second predictor variable in the model |
y |
response variable in the model |
scale |
logical; if |
normalize |
logical; if |
name.y1.hat |
Name for the vector corresponding to the marginal coefficient of |
name.y2.hat |
Name for the vector corresponding to the marginal coefficient of |
Details
If additional variables are included in the model, e.g., lm(y ~ x1 + x2 + x3 + ...), then
y, x1 and x2 are all taken as residuals from their separate linear fits
on x3 + ..., thus showing their partial relations net of (or adjusting for) these additional predictors.
A 3D diagram shows the vector y and the plane formed by the predictors,
x1 and x2, where all variables are represented in deviation form, so that
the intercept need not be included.
A 2D diagram, using the first two columns of the result, can be used to show the projection
of the space in the x1, x2 plane.
In these views, the ANOVA representation of the various sums of squares for the regression
predictors appears as the lengths of the various vectors. For example, the error sum of
squares is the squared length of the e vector, and the regression sum of squares is
the squared length of the yhat vector.
The drawing functions vectors and vectors3d used by the plot.regvec3d method only work
reasonably well if the variables are shown on commensurate scales, i.e., with
either scale=TRUE or normalize=TRUE.
Value
An object of class “regvec3d”, containing the following components
model |
The “lm” object corresponding to |
vectors |
A 9 x 3 matrix, whose rows correspond to the variables in the model,
the residual vector, the fitted vector, the partial fits for |
Methods (by class)
-
regvec3d(formula): Formula method for regvec3d -
regvec3d(default): Default method for regvec3d
References
Fox, J. (2016). Applied Regression Analysis and Generalized Linear Models, 3rd ed., Sage, Chapter 10.
Fox, J. and Friendly, M. (2016). "Visualizing Simultaneous Linear Equations, Geometric Vectors, and Least-Squares Regression with the matlib Package for R". useR Conference, Stanford, CA, June 27 - June 30, 2016.
See Also
Other vector diagrams:
Proj(),
arc(),
arrows3d(),
circle3d(),
corner(),
plot.regvec3d(),
pointOnLine(),
vectors(),
vectors3d()
Examples
library(rgl)
therapy.vec <- regvec3d(therapy ~ perstest + IE, data=therapy)
therapy.vec
plot(therapy.vec, col.plane="darkgreen")
plot(therapy.vec, dimension=2)
Row Cofactors of A[i,]
Description
Returns the vector of cofactors of row i of the square matrix A. The determinant, Det(A),
can then be found as M[i,] %*% rowCofactors(M,i) for any row, i.
Usage
rowCofactors(A, i)
Arguments
A |
a square matrix |
i |
row index |
Value
a vector of the cofactors of A[i,]
Author(s)
Michael Friendly
See Also
Det for the determinant
Other determinants:
Det(),
adjoint(),
cofactor(),
minor(),
rowMinors()
Examples
M <- matrix(c(4, -12, -4,
2, 1, 3,
-1, -3, 2), 3, 3, byrow=TRUE)
minor(M, 1, 1)
minor(M, 1, 2)
minor(M, 1, 3)
rowCofactors(M, 1)
Det(M)
# expansion by cofactors of row 1
M[1,] %*% rowCofactors(M,1)
Row Minors of A[i,]
Description
Returns the vector of minors of row i of the square matrix A
Usage
rowMinors(A, i)
Arguments
A |
a square matrix |
i |
row index |
Value
a vector of the minors of A[i,]
Author(s)
Michael Friendly
See Also
Other determinants:
Det(),
adjoint(),
cofactor(),
minor(),
rowCofactors()
Examples
M <- matrix(c(4, -12, -4,
2, 1, 3,
-1, -3, 2), 3, 3, byrow=TRUE)
minor(M, 1, 1)
minor(M, 1, 2)
minor(M, 1, 3)
rowMinors(M, 1)
Elementary Row Operations
Description
The elementary row operation rowadd adds multiples of one or more rows to other rows of a matrix.
This is usually used as a means to solve systems of linear equations, of the form A x = b, and rowadd
corresponds to adding equals to equals.
Usage
rowadd(x, from, to, mult)
Arguments
x |
a numeric matrix, possibly consisting of the coefficient matrix, A, joined with a vector of constants, b. |
from |
the index of one or more source rows. If |
to |
the index of one or more destination rows |
mult |
the multiplier(s) |
Details
The functions rowmult and rowswap complete the basic operations used in reduction
to row echelon form and Gaussian elimination. These functions are used for demonstration purposes.
Value
the matrix x, as modified
See Also
Other elementary row operations:
rowmult(),
rowswap()
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b <- c(8, -11, -3)
# using row operations to reduce below diagonal to 0
Ab <- cbind(A, b)
(Ab <- rowadd(Ab, 1, 2, 3/2)) # row 2 <- row 2 + 3/2 row 1
(Ab <- rowadd(Ab, 1, 3, 1)) # row 3 <- row 3 + 1 row 1
(Ab <- rowadd(Ab, 2, 3, -4)) # row 3 <- row 3 - 4 row 2
# multiply to make diagonals = 1
(Ab <- rowmult(Ab, 1:3, c(1/2, 2, -1)))
# The matrix is now in triangular form
# Could continue to reduce above diagonal to zero
echelon(A, b, verbose=TRUE, fractions=TRUE)
# convenient use of pipes
I <- diag( 3 )
AA <- I |>
rowadd(3, 1, 1) |> # add 1 x row 3 to row 1
rowadd(1, 3, 1) |> # add 1 x row 1 to row 3
rowmult(2, 2) # multiply row 2 by 2
Multiply Rows by Constants
Description
Multiplies one or more rows of a matrix by constants. This corresponds to multiplying or dividing equations by constants.
Usage
rowmult(x, row, mult)
Arguments
x |
a matrix, possibly consisting of the coefficient matrix, A, joined with a vector of constants, b. |
row |
index of one or more rows. |
mult |
row multiplier(s) |
Value
the matrix x, modified
See Also
Other elementary row operations:
rowadd(),
rowswap()
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b <- c(8, -11, -3)
# using row operations to reduce below diagonal to 0
Ab <- cbind(A, b)
(Ab <- rowadd(Ab, 1, 2, 3/2)) # row 2 <- row 2 + 3/2 row 1
(Ab <- rowadd(Ab, 1, 3, 1)) # row 3 <- row 3 + 1 row 1
(Ab <- rowadd(Ab, 2, 3, -4))
# multiply to make diagonals = 1
(Ab <- rowmult(Ab, 1:3, c(1/2, 2, -1)))
# The matrix is now in triangular form
Interchange two rows of a matrix
Description
This elementary row operation corresponds to interchanging two equations.
Usage
rowswap(x, from, to)
Arguments
x |
a matrix, possibly consisting of the coefficient matrix, A, joined with a vector of constants, b. |
from |
source row. |
to |
destination row |
Value
the matrix x, with rows from and to interchanged
See Also
Other elementary row operations:
rowadd(),
rowmult()
Show the eigenvectors associated with a covariance matrix
Description
This function is designed for illustrating the eigenvectors associated with the covariance matrix for a given bivariate data set. It draws a data ellipse of the data and adds vectors showing the eigenvectors of the covariance matrix.
Usage
showEig(
X,
col.vec = "blue",
lwd.vec = 3,
mult = sqrt(qchisq(levels, 2)),
asp = 1,
levels = c(0.5, 0.95),
plot.points = TRUE,
add = !plot.points,
...
)
Arguments
X |
A two-column matrix or data frame |
col.vec |
color for eigenvectors |
lwd.vec |
line width for eigenvectors |
mult |
length multiplier(s) for eigenvectors |
asp |
aspect ratio of plot, set to |
levels |
passed to dataEllipse determining the coverage of the data ellipse(s) |
plot.points |
logical; should the points be plotted? |
add |
logical; should this call add to an existing plot? |
... |
other arguments passed to |
Author(s)
Michael Friendly
See Also
Examples
x <- rnorm(200)
y <- .5 * x + .5 * rnorm(200)
X <- cbind(x,y)
showEig(X)
# Duncan data
data(Duncan, package="carData")
showEig(Duncan[, 2:3], levels=0.68)
showEig(Duncan[,2:3], levels=0.68, robust=TRUE, add=TRUE, fill=TRUE)
Show Matrices (A, b) as Linear Equations
Description
Shows what matrices \\mathbf{A}, \\mathbf{b} look like as the system of linear equations,
\\mathbf{A x} = \\mathbf{b}, but written out
as a set of equations.
Usage
showEqn(
A,
b,
vars,
simplify = FALSE,
reduce = FALSE,
fractions = FALSE,
latex = FALSE
)
Arguments
A |
either the matrix of coefficients of a system of linear equations, or the matrix |
b |
if supplied, the vector of constants on the right hand side of the equations. When omitted
the values |
vars |
a numeric or character vector of names of the variables.
If supplied, the length must be equal to the number of unknowns in the equations.
The default is |
simplify |
logical; try to simplify the equations? |
reduce |
logical; only show the unique linear equations |
fractions |
logical; express numbers as rational fractions, using the |
latex |
logical; print equations in a form suitable for LaTeX output? |
Value
a one-column character matrix, one row for each equation
Author(s)
Michael Friendly, John Fox, and Phil Chalmers
References
Fox, J. and Friendly, M. (2016). "Visualizing Simultaneous Linear Equations, Geometric Vectors, and Least-Squares Regression with the matlib Package for R". useR Conference, Stanford, CA, June 27 - June 30, 2016.
See Also
plotEqn, plotEqn3d, latexMatrix
Examples
A <- matrix(c(2, 1, -1,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
b <- c(8, -11, -3)
showEqn(A, b)
# show numerically
x <- solve(A, b)
showEqn(A, b, vars=x)
showEqn(A, b, simplify=TRUE)
showEqn(A, b, latex=TRUE)
# lower triangle of equation with zeros omitted (for back solving)
A <- matrix(c(2, 1, 2,
-3, -1, 2,
-2, 1, 2), 3, 3, byrow=TRUE)
U <- LU(A)$U
showEqn(U, simplify=TRUE, fractions=TRUE)
showEqn(U, b, simplify=TRUE, fractions=TRUE)
####################
# Linear models Design Matricies
data(mtcars)
ancova <- lm(mpg ~ wt + vs, mtcars)
summary(ancova)
showEqn(ancova)
showEqn(ancova, simplify=TRUE)
showEqn(ancova, vars=round(coef(ancova),2))
showEqn(ancova, vars=round(coef(ancova),2), simplify=TRUE)
twoway_int <- lm(mpg ~ vs * am, mtcars)
summary(twoway_int)
car::Anova(twoway_int)
showEqn(twoway_int)
showEqn(twoway_int, reduce=TRUE)
showEqn(twoway_int, reduce=TRUE, simplify=TRUE)
# Piece-wise linear regression
x <- c(1:10, 13:22)
y <- numeric(20)
y[1:10] <- 20:11 + rnorm(10, 0, 1.5)
y[11:20] <- seq(11, 15, len=10) + rnorm(10, 0, 1.5)
plot(x, y, pch = 16)
x2 <- as.numeric(x > 10)
mod <- lm(y ~ x + I((x - 10) * x2))
summary(mod)
lines(x, fitted(mod))
showEqn(mod)
showEqn(mod, vars=round(coef(mod),2))
showEqn(mod, simplify=TRUE)
Demonstrate the SVD for a 3 x 3 matrix
Description
This function draws an rgl scene consisting of a representation of the identity matrix and a
3 x 3 matrix A, together with the corresponding representation of the
matrices U, D, and V in the SVD decomposition,
A = U D V'.
Usage
svdDemo(A, shape = c("cube", "sphere"), alpha = 0.7, col = rainbow(6))
Arguments
A |
A 3 x 3 numeric matrix |
shape |
Basic shape used to represent the identity matrix: |
alpha |
transparency value used to draw the shape |
col |
Vector of 6 colors for the faces of the basic cube |
Value
Nothing
Author(s)
Original idea from Duncan Murdoch
Examples
A <- matrix(c(1,2,0.1, 0.1,1,0.1, 0.1,0.1,0.5), 3,3)
svdDemo(A)
## Not run:
B <- matrix(c( 1, 0, 1, 0, 2, 0, 1, 0, 2), 3, 3)
svdDemo(B)
# a positive, semi-definite matrix with eigenvalues 12, 6, 0
C <- matrix(c(7, 4, 1, 4, 4, 4, 1, 4, 7), 3, 3)
svdDemo(C)
## End(Not run)
The Matrix Sweep Operator
Description
The swp function “sweeps” a matrix on the rows and columns given in index to produce a new matrix
with those rows and columns “partialled out” by orthogonalization. This was defined as a fundamental statistical operation in
multivariate methods by Beaton (1964) and expanded by Dempster (1969). It is closely related to orthogonal projection,
but applied to a cross-products or covariance matrix, rather than to data.
Usage
swp(M, index)
Arguments
M |
a numeric matrix |
index |
a numeric vector indicating the rows/columns to be swept. The entries must be less than or equal
to the number or rows or columns in |
Details
If M is the partitioned matrix
\left[ \begin{array}{cc} \mathbf {R} & \mathbf {S} \\ \mathbf {T} & \mathbf {U} \end{array} \right]
where R is q \times q then swp(M, 1:q) gives
\left[ \begin{array}{cc} \mathbf {R}^{-1} & \mathbf {R}^{-1}\mathbf {S} \\ -\mathbf {TR}^{-1} & \mathbf {U}-\mathbf {TR}^{-1}\mathbf {S} \\ \end{array} \right]
Value
the matrix M with rows and columns in indices swept.
References
Beaton, A. E. (1964), The Use of Special Matrix Operations in Statistical Calculus, Princeton, NJ: Educational Testing Service.
Dempster, A. P. (1969) Elements of Continuous Multivariate Analysis. Addison-Wesley, Reading, Mass.
See Also
Examples
data(therapy)
mod3 <- lm(therapy ~ perstest + IE + sex, data=therapy)
X <- model.matrix(mod3)
XY <- cbind(X, therapy=therapy$therapy)
XY
M <- crossprod(XY)
swp(M, 1)
swp(M, 1:2)
Create a Symmetric Matrix from a Vector
Description
Creates a square symmetric matrix from a vector.
Usage
symMat(x, diag = TRUE, byrow = FALSE, names = FALSE)
Arguments
x |
A numeric vector used to fill the upper or lower triangle of the matrix. |
diag |
Logical. If |
byrow |
Logical. If |
names |
Either a logical or a character vector of names for the rows and columns of the matrix.
If |
Value
A symmetric square matrix based on column major ordering of the elements in x.
Author(s)
Originally from metaSEM::vec2symMat, Mike W.-L. Cheung <mikewlcheung@nus.edu.sg>; modified by Michael Friendly
Examples
symMat(1:6)
symMat(1:6, byrow=TRUE)
symMat(5:0, diag=FALSE)
Therapy Data
Description
A toy data set on outcome in therapy in relation to a personality test (perstest)
and a scale of internal-external locus of control (IE)
used to illustrate linear and multiple regression.
Usage
data("therapy")Format
A data frame with 10 observations on the following 4 variables.
sexa factor with levels
FMperstestscore on a personality test, a numeric vector
therapyoutcome in psychotherapy, a numeric vector
IEscore on a scale of internal-external locus of control, a numeric vector
Examples
data(therapy)
plot(therapy ~ perstest, data=therapy, pch=16)
abline(lm(therapy ~ perstest, data=therapy), col="red")
plot(therapy ~ perstest, data=therapy, cex=1.5, pch=16,
col=ifelse(sex=="M", "red","blue"))
Trace of a Matrix
Description
Calculates the trace of a square numeric matrix, i.e., the sum of its diagonal elements
Usage
tr(X)
Arguments
X |
a numeric matrix |
Value
a numeric value, the sum of diag(X)
Examples
X <- matrix(1:9, 3, 3)
tr(X)
Vandermode Matrix
Description
The function returns the Vandermode matrix of a numeric vector, x, whose columns are the vector
raised to the powers 0:n.
Usage
vandermode(x, n)
Arguments
x |
a numeric vector |
n |
a numeric scalar |
Value
a matrix of size length(x) x n
Examples
vandermode(1:5, 4)
Vectorize a Matrix
Description
Returns a 1-column matrix, stacking the columns of x, a matrix or vector.
Also supports comma-separated inputs similar to the concatenation
function c.
Usage
vec(x, ...)
Arguments
x |
A matrix or vector |
... |
(optional) additional objects to be stacked |
Value
A one-column matrix containing the elements of x and ...
in column order
Examples
vec(1:3)
vec(matrix(1:6, 2, 3))
vec(c("hello", "world"))
vec("hello", "world")
vec(1:3, "hello", "world")
Draw geometric vectors in 2D
Description
This function draws vectors in a 2D plot, in a way that facilitates constructing vector diagrams. It allows vectors to be specified as rows of a matrix, and can draw labels on the vectors.
Usage
vectors(
X,
origin = c(0, 0),
lwd = 2,
angle = 13,
length = 0.15,
labels = TRUE,
cex.lab = 1.5,
pos.lab = 4,
frac.lab = 1,
...
)
Arguments
X |
a vector or two-column matrix representing a set of geometric vectors; if a matrix, one vector is drawn for each row |
origin |
the origin from which they are drawn, a vector of length 2. |
lwd |
line width(s) for the vectors, a constant or vector of length equal to the number of rows of |
angle |
the |
length |
the |
labels |
a logical or a character vector of labels for the vectors. If |
cex.lab |
character expansion applied to vector labels. May be a number or numeric vector corresponding to the the
rows of |
pos.lab |
label position relative to the label point as in |
frac.lab |
location of label point, as a fraction of the distance between |
... |
other arguments passed on to graphics functions. |
Value
none
See Also
Other vector diagrams:
Proj(),
arc(),
arrows3d(),
circle3d(),
corner(),
plot.regvec3d(),
pointOnLine(),
regvec3d(),
vectors3d()
Examples
# shows addition of vectors
u <- c(3,1)
v <- c(1,3)
sum <- u+v
xlim <- c(0,5)
ylim <- c(0,5)
# proper geometry requires asp=1
plot( xlim, ylim, type="n", xlab="X", ylab="Y", asp=1)
abline(v=0, h=0, col="gray")
vectors(rbind(u,v,`u+v`=sum), col=c("red", "blue", "purple"), cex.lab=c(2, 2, 2.2))
# show the opposing sides of the parallelogram
vectors(sum, origin=u, col="red", lty=2)
vectors(sum, origin=v, col="blue", lty=2)
# projection of vectors
vectors(Proj(v,u), labels="P(v,u)", lwd=3)
vectors(v, origin=Proj(v,u))
corner(c(0,0), Proj(v,u), v, col="grey")
Draw 3D vectors
Description
This function draws vectors in a 3D plot, in a way that facilitates constructing vector diagrams. It allows vectors to be specified as rows of a matrix, and can draw labels on the vectors.
Usage
vectors3d(
X,
origin = c(0, 0, 0),
headlength = 0.035,
ref.length = NULL,
radius = 1/60,
labels = TRUE,
cex.lab = 1.2,
adj.lab = 0.5,
frac.lab = 1.1,
draw = TRUE,
...
)
Arguments
X |
a vector or three-column matrix representing a set of geometric vectors; if a matrix, one vector is drawn for each row |
origin |
the origin from which they are drawn, a vector of length 3. |
headlength |
the |
ref.length |
vector length to be used in scaling arrow heads so that they are all the same size; if |
radius |
radius of the base of the arrow heads |
labels |
a logical or a character vector of labels for the vectors. If |
cex.lab |
character expansion applied to vector labels. May be a number or numeric vector corresponding to the the
rows of |
adj.lab |
label position relative to the label point as in |
frac.lab |
location of label point, as a fraction of the distance between |
draw |
if |
... |
other arguments passed on to graphics functions. |
Value
invisibly returns the vector ref.length used to scale arrow heads
Bugs
At present, the color (color=) argument is not handled as expected when more than one vector is to be drawn.
Author(s)
Michael Friendly
See Also
arrows3d, texts3d, rgl.material
Other vector diagrams:
Proj(),
arc(),
arrows3d(),
circle3d(),
corner(),
plot.regvec3d(),
pointOnLine(),
regvec3d(),
vectors()
Examples
vec <- rbind(diag(3), c(1,1,1))
rownames(vec) <- c("X", "Y", "Z", "J")
library(rgl)
open3d()
vectors3d(vec, color=c(rep("black",3), "red"), lwd=2)
# draw the XZ plane, whose equation is Y=0
planes3d(0, 0, 1, 0, col="gray", alpha=0.2)
vectors3d(c(1,1,0), col="green", lwd=2)
# show projections of the unit vector J
segments3d(rbind(c(1,1,1), c(1, 1, 0)))
segments3d(rbind(c(0,0,0), c(1, 1, 0)))
segments3d(rbind(c(1,0,0), c(1, 1, 0)))
segments3d(rbind(c(0,1,0), c(1, 1, 0)))
# show some orthogonal vectors
p1 <- c(0,0,0)
p2 <- c(1,1,0)
p3 <- c(1,1,1)
p4 <- c(1,0,0)
corner(p1, p2, p3, col="red")
corner(p1, p4, p2, col="red")
corner(p1, p4, p3, col="blue")
rgl.bringtotop()
Workers Data
Description
A toy data set comprised of information on workers Income in relation
to other variables, used for illustrating linear and multiple regression.
Usage
data("workers")Format
A data frame with 10 observations on the following 4 variables.
Incomeincome from the job, a numeric vector
Experiencenumber of years of experience, a numeric vector
Skillskill level in the job, a numeric vector
Gendera factor with levels
FemaleMale
Examples
data(workers)
plot(Income ~ Experience, data=workers, main="Income ~ Experience", pch=20, cex=2)
# simple linear regression
reg1 <- lm(Income ~ Experience, data=workers)
abline(reg1, col="red", lwd=3)
# quadratic fit?
plot(Income ~ Experience, data=workers, main="Income ~ poly(Experience,2)", pch=20, cex=2)
reg2 <- lm(Income ~ poly(Experience,2), data=workers)
fit2 <-predict(reg2)
abline(reg1, col="red", lwd=1, lty=1)
lines(workers$Experience, fit2, col="blue", lwd=3)
# How does Income depend on a factor?
plot(Income ~ Gender, data=workers, main="Income ~ Gender")
points(workers$Gender, jitter(workers$Income), cex=2, pch=20)
means<-aggregate(workers$Income,list(workers$Gender),mean)
points(means,col="red", pch="+", cex=2)
lines(means,col="red", lwd=2)
Generalized Vector Cross Product
Description
Given two linearly independent length 3 vectors **a** and **b**, the cross product, \mathbf{a} \times \mathbf{b}
(read "a cross b"), is a vector that is perpendicular to both **a** and **b**
thus normal to the plane containing them.
Usage
xprod(...)
Arguments
... |
N-1 linearly independent vectors of the same length, N. |
Details
A generalization of this idea applies to two or more dimensional vectors.
See: [https://en.wikipedia.org/wiki/Cross_product] for geometric and algebraic properties.
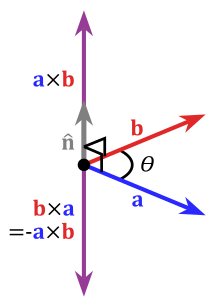
Value
Returns the generalized vector cross-product, a vector of length N.
Author(s)
Matthew Lundberg, in a [Stack Overflow post][https://stackoverflow.com/questions/36798301/r-compute-cross-product-of-vectors-physics]
Examples
xprod(1:3, 4:6)
# This works for an dimension
xprod(c(0,1)) # 2d
xprod(c(1,0,0), c(0,1,0)) # 3d
xprod(c(1,1,1), c(0,1,0)) # 3d
xprod(c(1,0,0,0), c(0,1,0,0), c(0,0,1,0)) # 4d